Evolutionary paths of streptococcal and staphylococcal superantigens
- PMID: 22900646
- PMCID: PMC3538662
- DOI: 10.1186/1471-2164-13-404
Evolutionary paths of streptococcal and staphylococcal superantigens
Abstract
Background: Streptococcus pyogenes (GAS) harbors several superantigens (SAgs) in the prophage region of its genome, although speG and smez are not located in this region. The diversity of SAgs is thought to arise during horizontal transfer, but their evolutionary pathways have not yet been determined. We recently completed sequencing the entire genome of S. dysgalactiae subsp. equisimilis (SDSE), the closest relative of GAS. Although speG is the only SAg gene of SDSE, speG was present in only 50% of clinical SDSE strains and smez in none. In this study, we analyzed the evolutionary paths of streptococcal and staphylococcal SAgs.
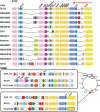
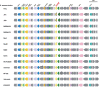
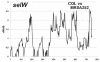
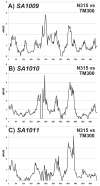
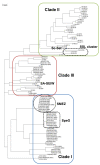
Results: We compared the sequences of the 12-60 kb speG regions of nine SDSE strains, five speG(+) and four speG(-). We found that the synteny of this region was highly conserved, whether or not the speG gene was present. Synteny analyses based on genome-wide comparisons of GAS and SDSE indicated that speG is the direct descendant of a common ancestor of streptococcal SAgs, whereas smez was deleted from SDSE after SDSE and GAS split from a common ancestor. Cumulative nucleotide skew analysis of SDSE genomes suggested that speG was located outside segments of steeper slopes than the stable region in the genome, whereas the region flanking smez was unstable, as expected from the results of GAS. We also detected a previously undescribed staphylococcal SAg gene, selW, and a staphylococcal SAg -like gene, ssl, in the core genomes of all Staphylococcus aureus strains sequenced. Amino acid substitution analyses, based on dN/dS window analysis of the products encoded by speG, selW and ssl suggested that all three genes have been subjected to strong positive selection. Evolutionary analysis based on the Bayesian Markov chain Monte Carlo method showed that each clade included at least one direct descendant.
Conclusions: Our findings reveal a plausible model for the comprehensive evolutionary pathway of streptococcal and staphylococcal SAgs.
Figures

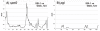




References
-
- Alouf JE, Müller H. In: The comprehensive sourcebook of bacterial protein toxins. 3. Alouf JE, Popoff MR, editor. New York: Academic Press; 2006. What are superantigens? pp. 821–829.
-
- Shimomura Y, Okumura K, Murayama SY, Yagi J, Ubukata K, Kirikae T, Miyoshi-Akiyama T. Complete genome sequencing and analysis of a Lancefield group G Streptococcus dysgalactiae subsp. equisimilis strain causing streptococcal toxic shock syndrome (STSS) BMC Genomics. 2011;12(1):17. doi: 10.1186/1471-2164-12-17. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

