Identification of a novel set of genes reflecting different in vivo invasive patterns of human GBM cells
- PMID: 22901239
- PMCID: PMC3502598
- DOI: 10.1186/1471-2407-12-358
Identification of a novel set of genes reflecting different in vivo invasive patterns of human GBM cells
Abstract
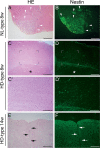
Background: Most patients affected by Glioblastoma multiforme (GBM, grade IV glioma) experience a recurrence of the disease because of the spreading of tumor cells beyond surgical boundaries. Unveiling mechanisms causing this process is a logic goal to impair the killing capacity of GBM cells by molecular targeting.We noticed that our long-term GBM cultures, established from different patients, may display two categories/types of growth behavior in an orthotopic xenograft model: expansion of the tumor mass and formation of tumor branches/nodules (nodular like, NL-type) or highly diffuse single tumor cell infiltration (HD-type).
Methods: We determined by DNA microarrays the gene expression profiles of three NL-type and three HD-type long-term GBM cultures. Subsequently, individual genes with different expression levels between the two groups were identified using Significance Analysis of Microarrays (SAM). Real time RT-PCR, immunofluorescence and immunoblot analyses, were performed for a selected subgroup of regulated gene products to confirm the results obtained by the expression analysis.
Results: Here, we report the identification of a set of 34 differentially expressed genes in the two types of GBM cultures. Twenty-three of these genes encode for proteins localized to the plasma membrane and 9 of these for proteins are involved in the process of cell adhesion.
Conclusions: This study suggests the participation in the diffuse infiltrative/invasive process of GBM cells within the CNS of a novel set of genes coding for membrane-associated proteins, which should be thus susceptible to an inhibition strategy by specific targeting.Massimiliano Monticone and Antonio Daga contributed equally to this work.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

