Cancer vulnerabilities unveiled by genomic loss
- PMID: 22901813
- PMCID: PMC3429351
- DOI: 10.1016/j.cell.2012.07.023
Cancer vulnerabilities unveiled by genomic loss
Abstract
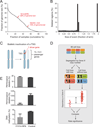
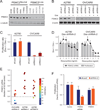
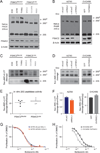
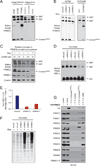
Due to genome instability, most cancers exhibit loss of regions containing tumor suppressor genes and collateral loss of other genes. To identify cancer-specific vulnerabilities that are the result of copy number losses, we performed integrated analyses of genome-wide copy number and RNAi profiles and identified 56 genes for which gene suppression specifically inhibited the proliferation of cells harboring partial copy number loss of that gene. These CYCLOPS (copy number alterations yielding cancer liabilities owing to partial loss) genes are enriched for spliceosome, proteasome, and ribosome components. One CYCLOPS gene, PSMC2, encodes an essential member of the 19S proteasome. Normal cells express excess PSMC2, which resides in a complex with PSMC1, PSMD2, and PSMD5 and acts as a reservoir protecting cells from PSMC2 suppression. Cells harboring partial PSMC2 copy number loss lack this complex and die after PSMC2 suppression. These observations define a distinct class of cancer-specific liabilities resulting from genome instability.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

References
-
- Aghajanian C, Soignet S, Dizon DS, Pien CS, Adams J, Elliott PJ, Sabbatini P, Miller V, Hensley ML, Pezzulli S, et al. A phase I trial of the novel proteasome inhibitor PS341 in advanced solid tumor malignancies. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:2505–2511. - PubMed
-
- Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B-Methodol. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

