The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E
- PMID: 22902561
- PMCID: PMC3469820
- DOI: 10.1016/j.molcel.2012.07.015
The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E
Abstract
Numerous small non-coding RNAs (sRNAs) in bacteria modulate rates of translation initiation and degradation of target mRNAs, which they recognize through base-pairing facilitated by the RNA chaperone Hfq. Recent evidence indicates that the ternary complex of Hfq, sRNA and mRNA guides endoribonuclease RNase E to initiate turnover of both the RNAs. We show that a sRNA not only guides RNase E to a defined site in a target RNA, but also allosterically activates the enzyme by presenting a monophosphate group at the 5'-end of the cognate-pairing "seed." Moreover, in the absence of the target the 5'-monophosphate makes the sRNA seed region vulnerable to an attack by RNase E against which Hfq confers no protection. These results suggest that the chemical signature and pairing status of the sRNA seed region may help to both 'proofread' recognition and activate mRNA cleavage, as part of a dynamic process involving cooperation of RNA, Hfq and RNase E.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




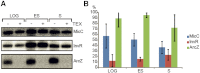
Comment in
-
RNase E finds some sRNAs stimulating.Mol Cell. 2012 Sep 28;47(6):825-6. doi: 10.1016/j.molcel.2012.09.007. Mol Cell. 2012. PMID: 23020853 Free PMC article.
References
-
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. - PubMed
-
- Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. - PubMed
-
- Balbontín R., Fiorini F., Figueroa-Bossi N., Casadesús J., Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 2010;78:380–394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

