Comparison of nerve growth factor receptor binding models using heterodimeric muteins
- PMID: 22903500
- PMCID: PMC3674885
- DOI: 10.1002/jnr.23116
Comparison of nerve growth factor receptor binding models using heterodimeric muteins
Abstract
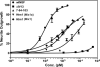
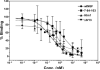
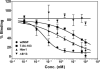
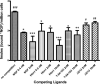
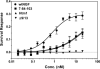
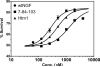
Nerve growth factor (NGF) is a homodimer that binds to two distinct receptor types, TrkA and p75, to support survival and differentiation of neurons. The high-affinity binding on the cell surface is believed to involve a heteroreceptor complex, but its exact nature is unclear. We developed a heterodimer (heteromutein) of two NGF muteins that can bind p75 and TrkA on opposite sides of the heterodimer, but not two TrkA receptors. Previously described muteins are Δ9/13 that is TrkA negative and 7-84-103 that is signal selective through TrkA. The heteromutein (Htm1) was used to study the heteroreceptor complex formation and function, in the putative absence of NGF-induced TrkA dimerization. Cellular binding assays indicated that Htm1 does not bind TrkA as efficiently as wild-type (wt) NGF but has better affinity than either homodimeric mutein. Htm1, 7-84-103, and Δ9/13 were each able to compete for cold-temperature, cold-chase stable binding on PC12 cells, indicating that binding to p75 was required for a portion of this high-affinity binding. Survival, neurite outgrowth, and MAPK signaling in PC12 cells also showed a reduced response for Htm1, compared with wtNGF, but was better than the parent muteins in the order wtNGF > Htm1 > 7-84-103 >> Δ9/13. Htm1 and 7-84-103 demonstrated similar levels of survival on cells expressing only TrkA. In the longstanding debate on the NGF receptor binding mechanism, our data support the ligand passing of NGF from p75 to TrkA involving a transient heteroreceptor complex of p75-NGF-TrkA.
Copyright © 2012 Wiley Periodicals, Inc.
Figures

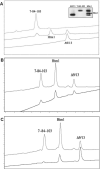

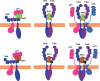
Similar articles
-
Identification of critical residues within the conserved and specificity patches of nerve growth factor leading to survival or differentiation.J Biol Chem. 2009 Nov 27;284(48):33600-13. doi: 10.1074/jbc.M109.058420. Epub 2009 Sep 17. J Biol Chem. 2009. PMID: 19762468 Free PMC article.
-
Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors.Neuron. 2007 Jan 4;53(1):25-38. doi: 10.1016/j.neuron.2006.09.034. Neuron. 2007. PMID: 17196528
-
Distinction between differentiation, cell cycle, and apoptosis signals in PC12 cells by the nerve growth factor mutant delta9/13, which is selective for the p75 neurotrophin receptor.J Neurosci Res. 2001 Jan 1;63(1):10-9. doi: 10.1002/1097-4547(20010101)63:1<10::AID-JNR2>3.0.CO;2-R. J Neurosci Res. 2001. PMID: 11169609
-
High affinity not in the vicinity?Neuron. 2007 Jan 4;53(1):1-4. doi: 10.1016/j.neuron.2006.12.018. Neuron. 2007. PMID: 17196523 Review.
-
Transcriptional down-regulation of epidermal growth factor (EGF) receptors by nerve growth factor (NGF) in PC12 cells.J Mol Neurosci. 2014 Nov;54(3):574-85. doi: 10.1007/s12031-014-0388-2. Epub 2014 Jul 31. J Mol Neurosci. 2014. PMID: 25078264 Review.
Cited by
-
Epistasis of HTR1A and BDNF risk genes alters cortical 5-HT1A receptor binding: PET results link genotype to molecular phenotype in depression.Transl Psychiatry. 2019 Jan 16;9(1):5. doi: 10.1038/s41398-018-0308-2. Transl Psychiatry. 2019. PMID: 30664620 Free PMC article.
-
Alternatively spliced, truncated GCSF receptor promotes leukemogenic properties and sensitivity to JAK inhibition.Leukemia. 2014 May;28(5):1041-51. doi: 10.1038/leu.2013.321. Epub 2013 Oct 30. Leukemia. 2014. PMID: 24170028 Free PMC article.
-
Unveiling functional motions based on point mutations in biased signaling systems: A normal mode study on nerve growth factor bound to TrkA.PLoS One. 2020 Jun 4;15(6):e0231542. doi: 10.1371/journal.pone.0231542. eCollection 2020. PLoS One. 2020. PMID: 32497034 Free PMC article.
-
The conundrum of the high-affinity NGF binding site formation unveiled?Biophys J. 2015 Feb 3;108(3):687-97. doi: 10.1016/j.bpj.2014.11.3485. Biophys J. 2015. PMID: 25650935 Free PMC article.
References
-
- Barker PA. High affinity not in the vicinity? Neuron. 2007;53:1–4. - PubMed
-
- Barker PA, Shooter EM. Disruption of NGF binding to the low affinity neurotrophin receptor p75LNTR reduces NGF binding to TrkA on PC12 cells. Neuron. 1994;13:203–215. - PubMed
-
- Barker PA, Lomen-Hoerth C, Gensch EM, Meakin SO, Glass DJ, Shooter EM. Tissue-specific alternative splicing generates two isoforms of the trkA receptor. J Biol Chem. 1993;268:15150–15157. - PubMed
-
- Barker PA, Hussain NK, McPherson PS. Retrograde signaling by the neurotrophins follows a well-worn trk. Trends Neurosci. 2002;25:379–381. - PubMed
-
- Bernd P, Greene LA. Association of 125I-nerve growth factor with PC12 pheochromocytoma cells. Evidence for internalization via high-affinity receptors only and for long-term regulation by nerve growth factor of both high- and low-affinity receptors. J Biol Chem. 1984;259:15509–15516. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

