Functional impact bias reveals cancer drivers
- PMID: 22904074
- PMCID: PMC3505979
- DOI: 10.1093/nar/gks743
Functional impact bias reveals cancer drivers
Abstract
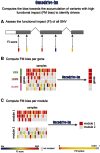
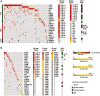
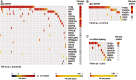
Identifying cancer driver genes and pathways among all somatic mutations detected in a cohort of tumors is a key challenge in cancer genomics. Traditionally, this is done by prioritizing genes according to the recurrence of alterations that they bear. However, this approach has some known limitations, such as the difficulty to correctly estimate the background mutation rate, and the fact that it cannot identify lowly recurrently mutated driver genes. Here we present a novel approach, Oncodrive-fm, to detect candidate cancer drivers which does not rely on recurrence. First, we hypothesized that any bias toward the accumulation of variants with high functional impact observed in a gene or group of genes may be an indication of positive selection and can thus be used to detect candidate driver genes or gene modules. Next, we developed a method to measure this bias (FM bias) and applied it to three datasets of tumor somatic variants. As a proof of concept of our hypothesis we show that most of the highly recurrent and well-known cancer genes exhibit a clear FM bias. Moreover, this novel approach avoids some known limitations of recurrence-based approaches, and can successfully identify lowly recurrent candidate cancer drivers.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

