Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets
- PMID: 22904076
- PMCID: PMC3479208
- DOI: 10.1093/nar/gks766
Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets
Abstract
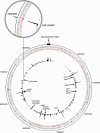
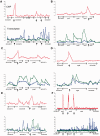
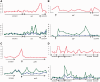
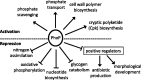
Streptomycetes sense and respond to the stress of phosphate starvation via the two-component PhoR-PhoP signal transduction system. To identify the in vivo targets of PhoP we have undertaken a chromatin-immunoprecipitation-on-microarray analysis of wild-type and phoP mutant cultures and, in parallel, have quantified their transcriptomes. Most (ca. 80%) of the previously in vitro characterized PhoP targets were identified in this study among several hundred other putative novel PhoP targets. In addition to activating genes for phosphate scavenging systems PhoP was shown to target two gene clusters for cell wall/extracellular polymer biosynthesis. Furthermore PhoP was found to repress an unprecedented range of pathways upon entering phosphate limitation including nitrogen assimilation, oxidative phosphorylation, nucleotide biosynthesis and glycogen catabolism. Moreover, PhoP was shown to target many key genes involved in antibiotic production and morphological differentiation, including afsS, atrA, bldA, bldC, bldD, bldK, bldM, cdaR, cdgA, cdgB and scbR-scbA. Intriguingly, in the PhoP-dependent cpk polyketide gene cluster, PhoP accumulates substantially at three specific sites within the giant polyketide synthase-encoding genes. This study suggests that, following phosphate limitation, Streptomyces coelicolor PhoP functions as a 'master' regulator, suppressing central metabolism, secondary metabolism and developmental pathways until sufficient phosphate is salvaged to support further growth and, ultimately, morphological development.
Figures

References
-
- Apel AK, Sola-Landa A, Rodriguez-Garcia A, Martin JF. Phosphate control of phoA, phoC and phoD gene expression in Streptomyces coelicolor reveals significant differences in binding of PhoP to their promoter regions. Microbiology. 2007;153:3527–3537. - PubMed
-
- Santos-Beneit F, Rodriguez-Garcia A, Apel AK, Martin JF. Phosphate and carbon source regulation of two PhoP-dependent glycerophosphodiester phosphodiesterase genes of Streptomyces coelicolor. Microbiology. 2009;155:1800–1811. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

