PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species
- PMID: 22904089
- PMCID: PMC3526259
- DOI: 10.1093/nar/gks757
PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species
Abstract
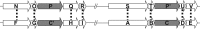
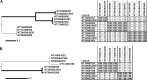
Pan-genome ortholog clustering tool (PanOCT) is a tool for pan-genomic analysis of closely related prokaryotic species or strains. PanOCT uses conserved gene neighborhood information to separate recently diverged paralogs into orthologous clusters where homology-only clustering methods cannot. The results from PanOCT and three commonly used graph-based ortholog-finding programs were compared using a set of four publicly available strains of the same bacterial species. All four methods agreed on ∼70% of the clusters and ∼86% of the proteins. The clusters that did not agree were inspected for evidence of correctness resulting in 85 high-confidence manually curated clusters that were used to compare all four methods.
Figures




References
-
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl Acad. Sci. USA. 2005;102:13950–13955. - PMC - PubMed
-
- Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. - PubMed
-
- Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. - PubMed
-
- Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–551. - PubMed

