Spg5 protein regulates the proteasome in quiescence
- PMID: 22904326
- PMCID: PMC3464545
- DOI: 10.1074/jbc.M112.390294
Spg5 protein regulates the proteasome in quiescence
Abstract
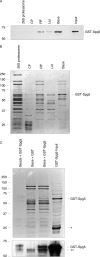
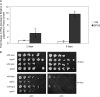
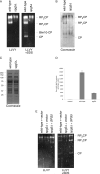
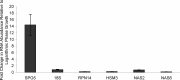
The ubiquitin-proteasome system is the major pathway for selective protein degradation in eukaryotes. Despite extensive study of this system, the mechanisms by which proteasome function and cell growth are coordinated remain unclear. Here, we identify Spg5 as a novel component of the ubiquitin-proteasome system. Spg5 binds the regulatory particle of the proteasome and the base subassembly in particular, but it is excluded from mature proteasomes. The SPG5 gene is strongly induced in the stationary phase of budding yeast, and spg5Δ mutants show a progressive loss of viability under these conditions. Accordingly, during logarithmic growth, Spg5 appears largely dispensable for proteasome function, but during stationary phase the proteasomes of spg5Δ mutants show both structural and functional defects. This loss of proteasome function is reflected in the accumulation of oxidized proteins preferentially in stationary phase in spg5Δ mutants. Thus, Spg5 is a positive regulator of the proteasome that is critical for survival of cells that have ceased to proliferate due to nutrient limitation.
Figures



References
-
- Teichert U., Mechler B., Müller H., Wolf D. H. (1989) Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J. Biol. Chem. 264, 16037–16045 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

