The Human Microbiome Project: a community resource for the healthy human microbiome
- PMID: 22904687
- PMCID: PMC3419203
- DOI: 10.1371/journal.pbio.1001377
The Human Microbiome Project: a community resource for the healthy human microbiome
Abstract
This manuscript describes the NIH Human Microbiome Project, including a brief review of human microbiome research, a history of the project, and a comprehensive overview of the consortium's recent collection of publications analyzing the human microbiome.
Conflict of interest statement
The authors have declared that no competing interests exist.
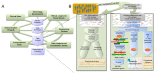
Figures
References
-
- Relman DA, Falkow S (2001) The meaning and impact of the human genome sequence for microbiology. Trends Microbiol 9: 206–208. - PubMed
-
- Davies J (2001) In a map for human life, count the microbes, too. Science 291: 2316. - PubMed
-
- Stahl DA, Lane DJ, Olsen GJ, Pace NR (1984) Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science 224: 409–411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 HG004973/HG/NHGRI NIH HHS/United States
- U54HG004969/HG/NHGRI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01HG004872/HG/NHGRI NIH HHS/United States
- U54 AI084844/AI/NIAID NIH HHS/United States
- U54HG004973/HG/NHGRI NIH HHS/United States
- R01 HG004872/HG/NHGRI NIH HHS/United States
- R01 HG005969/HG/NHGRI NIH HHS/United States
- U54 HG004969/HG/NHGRI NIH HHS/United States
- R01HG005969/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01HG004866/HG/NHGRI NIH HHS/United States
- U01 HG004866/HG/NHGRI NIH HHS/United States
- U54AI084844/AI/NIAID NIH HHS/United States

