Fascaplysin as a specific inhibitor for CDK4: insights from molecular modelling
- PMID: 22905154
- PMCID: PMC3419161
- DOI: 10.1371/journal.pone.0042612
Fascaplysin as a specific inhibitor for CDK4: insights from molecular modelling
Abstract
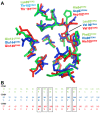
Cyclin-dependent kinases (CDKs) play a key role in the cell cycle and are important anti-cancer drug targets. The natural product fascaplysin inhibits CDK4 with surprising selectivity (IC(50) = 0.4 µM) compared to the close homolog CDK2 (IC(50) = 500 µM). Free energy calculations of the positively charged fascaplysin and an uncharged iso-electronic derivative in the CDK2 and CDK4 inhibitor complexes indicate that the positive charge of fascaplysin is crucial for selectivity. This finding will guide further improvements in the design of fascaplysin-based selective inhibitors for CDK4.
Conflict of interest statement
Figures




Similar articles
-
Inhibition of cyclin-dependent kinase 4 (Cdk4) by fascaplysin, a marine natural product.Biochem Biophys Res Commun. 2000 Sep 7;275(3):877-84. doi: 10.1006/bbrc.2000.3349. Biochem Biophys Res Commun. 2000. PMID: 10973815
-
CA224, a non-planar analogue of fascaplysin, inhibits Cdk4 but not Cdk2 and arrests cells at G0/G1 inhibiting pRB phosphorylation.Bioorg Med Chem Lett. 2006 Aug 15;16(16):4272-8. doi: 10.1016/j.bmcl.2006.05.065. Epub 2006 Jun 5. Bioorg Med Chem Lett. 2006. PMID: 16750360
-
Design, synthesis and biological activity of new CDK4-specific inhibitors, based on fascaplysin.Org Biomol Chem. 2006 Mar 7;4(5):787-801. doi: 10.1039/b518019h. Epub 2006 Feb 1. Org Biomol Chem. 2006. PMID: 16493461
-
Recent research in selective cyclin-dependent kinase 4 inhibitors for anti-cancer treatment.Curr Med Chem. 2009;16(36):4869-88. doi: 10.2174/092986709789909611. Curr Med Chem. 2009. PMID: 19929781 Review.
-
Structure-based discovery and optimization of potential cancer therapeutics targeting the cell cycle.IDrugs. 2006 Apr;9(4):273-8. IDrugs. 2006. PMID: 16596481 Review.
Cited by
-
Antitumour potential of BPT: a dual inhibitor of cdk4 and tubulin polymerization.Cell Death Dis. 2015 May 7;6(5):e1743. doi: 10.1038/cddis.2015.96. Cell Death Dis. 2015. PMID: 25950473 Free PMC article.
-
Selective CDK9 Inhibition by Natural Compound Toyocamycin in Cancer Cells.Cancers (Basel). 2022 Jul 8;14(14):3340. doi: 10.3390/cancers14143340. Cancers (Basel). 2022. PMID: 35884401 Free PMC article.
-
Fascaplysin Sensitizes Anti-Cancer Effects of Drugs Targeting AKT and AMPK.Molecules. 2017 Dec 24;23(1):42. doi: 10.3390/molecules23010042. Molecules. 2017. PMID: 29295560 Free PMC article.
-
Marine-derived protein kinase inhibitors for neuroinflammatory diseases.Biomed Eng Online. 2018 Apr 24;17(1):46. doi: 10.1186/s12938-018-0477-5. Biomed Eng Online. 2018. PMID: 29690896 Free PMC article. Review.
-
Multiomics Analysis of Spatially Distinct Stromal Cells Reveals Tumor-Induced O-Glycosylation of the CDK4-pRB Axis in Fibroblasts at the Invasive Tumor Edge.Cancer Res. 2022 Feb 15;82(4):648-664. doi: 10.1158/0008-5472.CAN-21-1705. Cancer Res. 2022. PMID: 34853070 Free PMC article.
References
-
- Harper JW, Adams PD (2001) Cyclin-dependent kinases. Chem Rev 101: 2511–2526. - PubMed
-
- Morgan D (1997) Cyclin-dependent kinases: Engines, Clocks, and Microprocessors. Annual Review of Cell and Developmental Biology 13: 261–291. - PubMed
-
- Morgan DO (1995) Principles of CDK regulation. Nature 374: 131–134. - PubMed
-
- Norbury C, Nurse P (1992) Animal cell cycles and their control. Annual Review of Biochemistry 61: 441–468. - PubMed
-
- Malumbres M, Barbacid M (2005) Mammalian cyclin-dependent kinases. Trends in biochemical sciences 30: 630–641. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

