Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome
- PMID: 22905200
- PMCID: PMC3419686
- DOI: 10.1371/journal.pone.0043052
Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome
Abstract
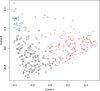
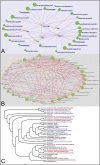
Obesity has been linked to the human gut microbiota; however, the contribution of gut bacterial species to the obese phenotype remains controversial because of conflicting results from studies in different populations. To explore the possible dysbiosis of gut microbiota in obesity and its metabolic complications, we studied men and women over a range of body mass indices from the Old Order Amish sect, a culturally homogeneous Caucasian population of Central European ancestry. We characterized the gut microbiota in 310 subjects by deep pyrosequencing of bar-coded PCR amplicons from the V1-V3 region of the 16S rRNA gene. Three communities of interacting bacteria were identified in the gut microbiota, analogous to previously identified gut enterotypes. Neither BMI nor any metabolic syndrome trait was associated with a particular gut community. Network analysis identified twenty-two bacterial species and four OTUs that were either positively or inversely correlated with metabolic syndrome traits, suggesting that certain members of the gut microbiota may play a role in these metabolic derangements.
Conflict of interest statement
Figures

References
-
- Mokdad AH, Marks JS, Stroup DF, Gerberding JL (2004) Actual causes of death in the United States. JAMA 291: 238–1245. - PubMed
-
- Power ML, Schulkin J (2008) Sex differences in fat storage, fat metabolism, and the health risks from obesity: possible evolutionary origins. Br. J. Nutrition 99: 931–940. - PubMed
-
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031 (2006).. - PubMed

