Nuclear organization and genome function
- PMID: 22905954
- PMCID: PMC3717390
- DOI: 10.1146/annurev-cellbio-101011-155824
Nuclear organization and genome function
Abstract
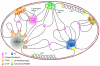
Long-range interactions between transcription regulatory elements play an important role in gene activation, epigenetic silencing, and chromatin organization. Transcriptional activation or repression of developmentally regulated genes is often accomplished through tissue-specific chromatin architecture and dynamic localization between active transcription factories and repressive Polycomb bodies. However, the mechanisms underlying the structural organization of chromatin and the coordination of physical interactions are not fully understood. Insulators and Polycomb group proteins form highly conserved multiprotein complexes that mediate functional long-range interactions and have proposed roles in nuclear organization. In this review, we explore recent findings that have broadened our understanding of the function of these proteins and provide an integrative model for the roles of insulators in nuclear organization.
Figures

References
-
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. - PubMed
-
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–26. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

