The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite
- PMID: 22906166
- PMCID: PMC3491370
- DOI: 10.1186/gb-2012-13-8-r70
The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite
Abstract
Background: The X-linked macrosatellite DXZ4 is a large homogenous tandem repeat that in females adopts an alternative chromatin organization on the primate X chromosome in response to X-chromosome inactivation. It is packaged into heterochromatin on the active X chromosome but into euchromatin and bound by the epigenetic organizer protein CTCF on the inactive X chromosome. Because its DNA sequence diverges rapidly beyond the New World monkeys, the existence of DXZ4 outside the primate lineage is unknown.
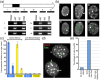
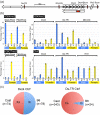
Results: Here we extend our comparative genome analysis and report the identification and characterization of the mouse homolog of the macrosatellite. Furthermore, we provide evidence of DXZ4 in a conserved location downstream of the PLS3 gene in a diverse group of mammals, and reveal that DNA sequence conservation is restricted to the CTCF binding motif, supporting a central role for this protein at this locus. However, many features that characterize primate DXZ4 differ in mouse, including the overall size of the array, the mode of transcription, the chromatin organization and conservation between adjacent repeat units of DNA sequence and length. Ctcf binds Dxz4 but is not exclusive to the inactive X chromosome, as evidenced by association in some males and equal binding to both X chromosomes in trophoblast stem cells.
Conclusions: Characterization of Dxz4 reveals substantial differences in the organization of DNA sequence, chromatin packaging, and the mode of transcription, so the potential roles performed by this sequence in mouse have probably diverged from those on the primate X chromosome.
Figures





References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. - DOI - PubMed
-
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

