Sirtuins of parasitic protozoa: in search of function(s)
- PMID: 22906508
- PMCID: PMC3484402
- DOI: 10.1016/j.molbiopara.2012.08.003
Sirtuins of parasitic protozoa: in search of function(s)
Abstract
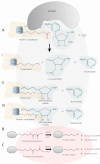
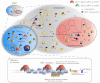
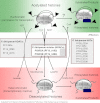
The SIR2 family of NAD(+)-dependent protein deacetylases, collectively called sirtuins, has been of central interest due to their proposed roles in life-span regulation and ageing. Sirtuins are one group of environment sensors of a cell interpreting external information and orchestrating internal responses at the sub-cellular level, through participation in gene regulation mechanisms. Remarkably conserved across all kingdoms of life SIR2 proteins in several protozoan parasites appear to have both conserved and intriguing unique functions. This review summarises our current knowledge of the members of the sirtuin families in Apicomplexa, including Plasmodium, and other protozoan parasites such as Trypanosoma and Leishmania. The wide diversity of processes regulated by SIR2 proteins makes them targets worthy of exploitation in anti-parasitic therapies.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

