Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation
- PMID: 22907758
- PMCID: PMC3486150
- DOI: 10.1128/MCB.06785-11
Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation
Abstract
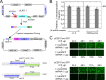
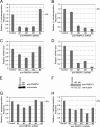
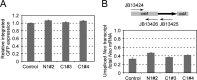
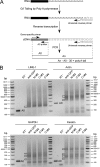
Poly(A) binding proteins (PABPs) specifically bind the polyadenosine tail of mRNA and have been shown to be important for RNA polyadenylation, translation initiation, and mRNA stability. Using a modified L1 retrotransposition vector, we examined the effects of two PABPs (encoded by PABPN1 and PABPC1) on the retrotransposition activity of the L1 non-long-terminal-repeat (non-LTR) retrotransposon in both HeLa and HEK293T cells. We demonstrated that knockdown of these two genes by RNA interference (RNAi) effectively reduced L1 retrotransposition by 70 to 80% without significantly changing L1 transcription or translation or the status of the poly(A) tail. We identified that both poly(A) binding proteins were associated with the L1 ribonucleoprotein complex, presumably through L1 mRNA. Depletion of PABPC1 caused a defect in L1 RNP formation. Knockdown of the PABPC1 inhibitor PAIP2 increased L1 retrotransposition up to 2-fold. Low levels of exogenous overexpression of PABPN1 and PABPC1 increased L1 retrotransposition, whereas unregulated overexpression of these two proteins caused pleiotropic effects, such as hypersensitivity to puromycin and decreased L1 activity. Our data suggest that PABPC1 is essential for the formation of L1 RNA-protein complexes and may play a role in L1 RNP translocation in the host cell.
Figures


Similar articles
-
Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions.Wiley Interdiscip Rev RNA. 2014 Sep-Oct;5(5):601-22. doi: 10.1002/wrna.1233. Epub 2014 Apr 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24789627 Free PMC article. Review.
-
hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition.Nucleic Acids Res. 2013 Jan 7;41(1):575-85. doi: 10.1093/nar/gks1075. Epub 2012 Nov 17. Nucleic Acids Res. 2013. PMID: 23161687 Free PMC article.
-
Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition.Hum Mol Genet. 2005 Nov 1;14(21):3237-48. doi: 10.1093/hmg/ddi354. Epub 2005 Sep 23. Hum Mol Genet. 2005. PMID: 16183655
-
Cytoplasmic poly(A)-binding protein 1 (PABPC1) interacts with the RNA-binding protein hnRNPLL and thereby regulates immunoglobulin secretion in plasma cells.J Biol Chem. 2017 Jul 21;292(29):12285-12295. doi: 10.1074/jbc.M117.794834. Epub 2017 Jun 13. J Biol Chem. 2017. PMID: 28611064 Free PMC article.
-
Crossing the borders: poly(A)-binding proteins working on both sides of the fence.RNA Biol. 2010 May-Jun;7(3):291-5. doi: 10.4161/rna.7.3.11649. Epub 2010 May 27. RNA Biol. 2010. PMID: 20400847 Review.
Cited by
-
Affinity Purification of the RNA Degradation Complex, the Exosome, from HEK-293 Cells.Bio Protoc. 2017 Apr 20;7(8):e2238. doi: 10.21769/BioProtoc.2238. Bio Protoc. 2017. PMID: 28691041 Free PMC article.
-
A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors.Nat Methods. 2018 May;15(5):330-338. doi: 10.1038/nmeth.4632. Epub 2018 Mar 19. Nat Methods. 2018. PMID: 29638227 Free PMC article.
-
Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line.Curr Issues Mol Biol. 2022 Feb 25;44(3):1115-1126. doi: 10.3390/cimb44030073. Curr Issues Mol Biol. 2022. PMID: 35723296 Free PMC article.
-
BRCA1 and S phase DNA repair pathways restrict LINE-1 retrotransposition in human cells.Nat Struct Mol Biol. 2020 Feb;27(2):179-191. doi: 10.1038/s41594-020-0374-z. Epub 2020 Feb 10. Nat Struct Mol Biol. 2020. PMID: 32042152 Free PMC article.
-
Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions.Wiley Interdiscip Rev RNA. 2014 Sep-Oct;5(5):601-22. doi: 10.1002/wrna.1233. Epub 2014 Apr 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24789627 Free PMC article. Review.
References
-
- Afonina E, Stauber R, Pavlakis GN. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273: 13015–13021 - PubMed
-
- An W, et al. 2011. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob. DNA 2: 2 doi:10.1186/1759-8753-2-2 - DOI - PMC - PubMed
-
- Athanikar JN, Badge RM, Moran JV. 2004. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 32: 3846–3855 doi:10.1093/nar/gkh698 - DOI - PMC - PubMed
-
- Belancio VP, Whelton M, Deininger P. 2007. Requirements for polyadenylation at the 3′ end of LINE-1 elements. Gene 390: 98–107 doi:10.1016/j.gene.2006.07.029 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
