GLISTR: glioma image segmentation and registration
- PMID: 22907965
- PMCID: PMC4371551
- DOI: 10.1109/TMI.2012.2210558
GLISTR: glioma image segmentation and registration
Abstract
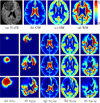
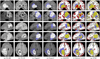
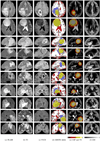
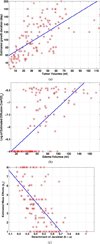
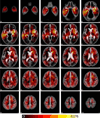
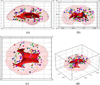
We present a generative approach for simultaneously registering a probabilistic atlas of a healthy population to brain magnetic resonance (MR) scans showing glioma and segmenting the scans into tumor as well as healthy tissue labels. The proposed method is based on the expectation maximization (EM) algorithm that incorporates a glioma growth model for atlas seeding, a process which modifies the original atlas into one with tumor and edema adapted to best match a given set of patient's images. The modified atlas is registered into the patient space and utilized for estimating the posterior probabilities of various tissue labels. EM iteratively refines the estimates of the posterior probabilities of tissue labels, the deformation field and the tumor growth model parameters. Hence, in addition to segmentation, the proposed method results in atlas registration and a low-dimensional description of the patient scans through estimation of tumor model parameters. We validate the method by automatically segmenting 10 MR scans and comparing the results to those produced by clinical experts and two state-of-the-art methods. The resulting segmentations of tumor and edema outperform the results of the reference methods, and achieve a similar accuracy from a second human rater. We additionally apply the method to 122 patients scans and report the estimated tumor model parameters and their relations with segmentation and registration results. Based on the results from this patient population, we construct a statistical atlas of the glioma by inverting the estimated deformation fields to warp the tumor segmentations of patients scans into a common space.
Figures




References
-
- Collins V. Gliomas. Cancer Survey. 1998;32:37–51. - PubMed
-
- Markert J, Devita V, Rosenberg S, Hellman S. Gliobalastoma Multiforme. Burlington, MA: Jones Bartlett; 2005.
-
- Weizman L, Sira L, Joskowicz L, Constantini S, Precel R, Shortly B, Bashat D. Automatic segmentation, internal classification, and follow-up of optic pathway gliomas in MRI. Med. Image Anal. 2012;16:177–188. - PubMed
-
- Liu J, Udupa J, Odhner D, Hackney D, Moonis G. A system for brain tumor volume estimation via MR imaging and fuzzy connectedness. Comput. Med. Imag. Graphics. 2005;29:21–34. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

