Genome-wide association mapping in tomato (Solanum lycopersicum) is possible using genome admixture of Solanum lycopersicum var. cerasiforme
- PMID: 22908034
- PMCID: PMC3411241
- DOI: 10.1534/g3.112.002667
Genome-wide association mapping in tomato (Solanum lycopersicum) is possible using genome admixture of Solanum lycopersicum var. cerasiforme
Abstract
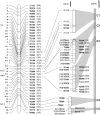
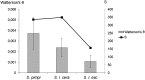
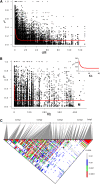
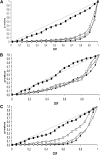
Genome-wide association mapping is an efficient way to identify quantitative trait loci controlling the variation of phenotypes, but the approach suffers severe limitations when one is studying inbred crops like cultivated tomato (Solanum lycopersicum). Such crops exhibit low rates of molecular polymorphism and high linkage disequilibrium, which reduces mapping resolution. The cherry type tomato (S. lycopersicum var. cerasiforme) genome has been described as an admixture between the cultivated tomato and its wild ancestor, S. pimpinellifolium. We have thus taken advantage of the properties of this admixture to improve the resolution of association mapping in tomato. As a proof of concept, we sequenced 81 DNA fragments distributed on chromosome 2 at different distances in a core collection of 90 tomato accessions, including mostly cherry type tomato accessions. The 81 Sequence Tag Sites revealed 352 SNPs and indels. Molecular diversity was greatest for S. pimpinellifolium accessions, intermediate for S. l. cerasiforme accessions, and lowest for the cultivated group. We assessed the structure of molecular polymorphism and the extent of linkage disequilibrium over genetic and physical distances. Linkage disequilibrium decreased under r(2) = 0.3 within 1 cM, and minimal estimated value (r(2) = 0.13) was reached within 20 kb over the physical regions studied. Associations between polymorphisms and fruit weight, locule number, and soluble solid content were detected. Several candidate genes and quantitative trait loci previously identified were validated and new associations detected. This study shows the advantages of using a collection of S. l. cerasiforme accessions to overcome the low resolution of association mapping in tomato.
Keywords: admixture; association mapping; linkage disequilibrium; tomato (Solanum lycopersicum).
Figures
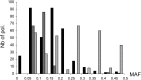

References
-
- Ardlie K. G., Kruglyak L., Seielstad M., 2002. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3: 299–309 - PubMed
-
- Benjamini Y., Hochberg Y., 2000. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25: 60–83
-
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., et al. , 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 - PubMed
-
- Buckler I., Edward S., Thornsberry J. M., 2002. Plant molecular diversity and applications to genomics. Curr. Opin. Plant Biol. 5: 107–111 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
