The UCSC genome browser and associated tools
- PMID: 22908213
- PMCID: PMC3603215
- DOI: 10.1093/bib/bbs038
The UCSC genome browser and associated tools
Abstract
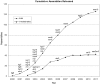
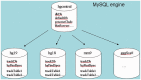
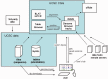
The UCSC Genome Browser (http://genome.ucsc.edu) is a graphical viewer for genomic data now in its 13th year. Since the early days of the Human Genome Project, it has presented an integrated view of genomic data of many kinds. Now home to assemblies for 58 organisms, the Browser presents visualization of annotations mapped to genomic coordinates. The ability to juxtapose annotations of many types facilitates inquiry-driven data mining. Gene predictions, mRNA alignments, epigenomic data from the ENCODE project, conservation scores from vertebrate whole-genome alignments and variation data may be viewed at any scale from a single base to an entire chromosome. The Browser also includes many other widely used tools, including BLAT, which is useful for alignments from high-throughput sequencing experiments. Private data uploaded as Custom Tracks and Data Hubs in many formats may be displayed alongside the rich compendium of precomputed data in the UCSC database. The Table Browser is a full-featured graphical interface, which allows querying, filtering and intersection of data tables. The Saved Session feature allows users to store and share customized views, enhancing the utility of the system for organizing multiple trains of thought. Binary Alignment/Map (BAM), Variant Call Format and the Personal Genome Single Nucleotide Polymorphisms (SNPs) data formats are useful for visualizing a large sequencing experiment (whole-genome or whole-exome), where the differences between the data set and the reference assembly may be displayed graphically. Support for high-throughput sequencing extends to compact, indexed data formats, such as BAM, bigBed and bigWig, allowing rapid visualization of large datasets from RNA-seq and ChIP-seq experiments via local hosting.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

