Breathing fluctuations in position-specific DNA base pairs are involved in regulating helicase movement into the replication fork
- PMID: 22908246
- PMCID: PMC3437866
- DOI: 10.1073/pnas.1212929109
Breathing fluctuations in position-specific DNA base pairs are involved in regulating helicase movement into the replication fork
Abstract
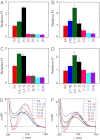
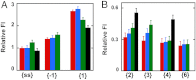
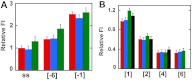
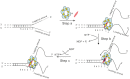
We previously used changes in the near-UV circular dichroism and fluorescence spectra of DNA base analogue probes placed site specifically to show that the first three base pairs at the fork junction in model replication fork constructs are significantly opened by "breathing" fluctuations under physiological conditions. Here, we use these probes to provide mechanistic snapshots of the initial interactions of the DNA fork with a tight-binding replication helicase in solution. The primosome helicase of bacteriophage T4 was assembled from six (gp41) helicase subunits, one (gp61) primase subunit, and nonhydrolyzable GTPγS. When bound to a DNA replication fork construct this complex advances one base pair into the duplex portion of the fork and forms a stably bound helicase "initiation complex." Replacement of GTPγS with GTP permits the completion of the helicase-driven unwinding process. Our spectroscopic probes show that the primosome in this stable helicase initiation complex binds the DNA of the fork primarily via backbone contacts and holds the first complementary base pair of the fork in an open conformation, whereas the second, third, and fourth base pairs of the duplex show essentially the breathing behavior that previously characterized the first three base pairs of the free fork. These spectral changes, together with dynamic fluorescence quenching results, are consistent with a primosome-binding model in which the lagging DNA strand passes through the central hole of the hexagonal helicase, the leading strand binds to the "outside" surfaces of subunits of the helicase hexamer, and the single primase subunit interacts with both strands.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. - PubMed
-
- Lohman TM. Escherichia coli DNA helicases: Mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. - PubMed
-
- Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

