Ribosomal protein S1 unwinds double-stranded RNA in multiple steps
- PMID: 22908248
- PMCID: PMC3437903
- DOI: 10.1073/pnas.1208950109
Ribosomal protein S1 unwinds double-stranded RNA in multiple steps
Abstract
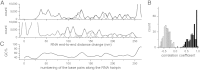
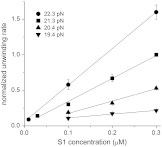
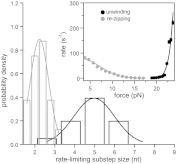
The sequence and secondary structure of the 5'-end of mRNAs regulate translation by controlling ribosome initiation on the mRNA. Ribosomal protein S1 is crucial for ribosome initiation on many natural mRNAs, particularly for those with structured 5'-ends, or with no or weak Shine-Dalgarno sequences. Besides a critical role in translation, S1 has been implicated in several other cellular processes, such as transcription recycling, and the rescuing of stalled ribosomes by tmRNA. The mechanisms of S1 functions are still elusive but have been widely considered to be linked to the affinity of S1 for single-stranded RNA and its corresponding destabilization of mRNA secondary structures. Here, using optical tweezers techniques, we demonstrate that S1 promotes RNA unwinding by binding to the single-stranded RNA formed transiently during the thermal breathing of the RNA base pairs and that S1 dissociation results in RNA rezipping. We measured the dependence of the RNA unwinding and rezipping rates on S1 concentration, and the force applied to the ends of the RNA. We found that each S1 binds 10 nucleotides of RNA in a multistep fashion implying that S1 can facilitate ribosome initiation on structured mRNA by first binding to the single strand next to an RNA duplex structure ("stand-by site") before subsequent binding leads to RNA unwinding. Unwinding by multiple small substeps is much less rate limited by thermal breathing than unwinding in a single step. Thus, a multistep scheme greatly expedites S1 unwinding of an RNA structure compared to a single-step mode.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

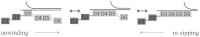
References
-
- Draper DE, von Hippel PH. Nucleic acid binding properties of Escherichia coli ribosomal protein S1. I. Structure and interactions of binding site I. J Mol Biol. 1978;122:321–338. - PubMed
-
- Subramanian AR, van Duin J. Exchange of individual ribosomal proteins between ribosomes as studied by heavy isotope-transfer experiments. Mol Gen Genet. 1977;158:1–9. - PubMed
-
- Marzi S, et al. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007;130:1019–1031. - PubMed
-
- Studer SM, Joseph S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell. 2006;22:105–115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

