Hsp110 is required for spindle length control
- PMID: 22908312
- PMCID: PMC3514029
- DOI: 10.1083/jcb.201111105
Hsp110 is required for spindle length control
Abstract
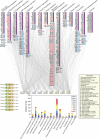
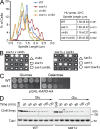
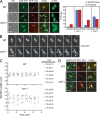
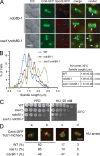
Systematic affinity purification combined with mass spectrometry analysis of N- and C-tagged cytoplasmic Hsp70/Hsp110 chaperones was used to identify new roles of Hsp70/Hsp110 in the cell. This allowed the mapping of a chaperone-protein network consisting of 1,227 unique interactions between the 9 chaperones and 473 proteins and highlighted roles for Hsp70/Hsp110 in 14 broad biological processes. Using this information, we uncovered an essential role for Hsp110 in spindle assembly and, more specifically, in modulating the activity of the widely conserved kinesin-5 motor Cin8. The role of Hsp110 Sse1 as a nucleotide exchange factor for the Hsp70 chaperones Ssa1/Ssa2 was found to be required for maintaining the proper distribution of kinesin-5 motors within the spindle, which was subsequently required for bipolar spindle assembly in S phase. These data suggest a model whereby the Hsp70-Hsp110 chaperone complex antagonizes Cin8 plus-end motility and prevents premature spindle elongation in S phase.
Figures




Similar articles
-
The control of spindle length by Hsp70 and Hsp110 molecular chaperones.FEBS Lett. 2013 Apr 17;587(8):1067-72. doi: 10.1016/j.febslet.2013.02.018. Epub 2013 Feb 19. FEBS Lett. 2013. PMID: 23434584 Review.
-
The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb.J Biol Chem. 2005 Dec 16;280(50):41262-9. doi: 10.1074/jbc.M503614200. Epub 2005 Oct 12. J Biol Chem. 2005. PMID: 16221677
-
Hsp110 is a nucleotide-activated exchange factor for Hsp70.J Biol Chem. 2008 Apr 4;283(14):8877-84. doi: 10.1074/jbc.M710063200. Epub 2008 Jan 24. J Biol Chem. 2008. PMID: 18218635
-
Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1.Biochemistry. 2006 Dec 19;45(50):15075-84. doi: 10.1021/bi061279k. Biochemistry. 2006. PMID: 17154545 Free PMC article.
-
GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones.Subcell Biochem. 2015;78:1-33. doi: 10.1007/978-3-319-11731-7_1. Subcell Biochem. 2015. PMID: 25487014 Review.
Cited by
-
A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane.PLoS Pathog. 2015 Aug 5;11(8):e1005086. doi: 10.1371/journal.ppat.1005086. eCollection 2015 Aug. PLoS Pathog. 2015. PMID: 26244546 Free PMC article.
-
CBP-HSF2 structural and functional interplay in Rubinstein-Taybi neurodevelopmental disorder.Nat Commun. 2022 Nov 16;13(1):7002. doi: 10.1038/s41467-022-34476-2. Nat Commun. 2022. PMID: 36385105 Free PMC article.
-
HSP70 regulates the function of mitotic centrosomes.Cell Mol Life Sci. 2016 Oct;73(20):3949-60. doi: 10.1007/s00018-016-2236-8. Epub 2016 Apr 30. Cell Mol Life Sci. 2016. PMID: 27137183 Free PMC article.
-
Coordinated Hsp110 and Hsp104 Activities Power Protein Disaggregation in Saccharomyces cerevisiae.Mol Cell Biol. 2017 May 16;37(11):e00027-17. doi: 10.1128/MCB.00027-17. Print 2017 Jun 1. Mol Cell Biol. 2017. PMID: 28289075 Free PMC article.
-
Condensin ATPase motifs contribute differentially to the maintenance of chromosome morphology and genome stability.PLoS Biol. 2018 Jun 27;16(6):e2003980. doi: 10.1371/journal.pbio.2003980. eCollection 2018 Jun. PLoS Biol. 2018. PMID: 29949571 Free PMC article.
References
-
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

