Direct vs 2-stage approaches to structured motif finding
- PMID: 22908910
- PMCID: PMC3564690
- DOI: 10.1186/1748-7188-7-20
Direct vs 2-stage approaches to structured motif finding
Abstract
Background: The notion of DNA motif is a mathematical abstraction used to model regions of the DNA (known as Transcription Factor Binding Sites, or TFBSs) that are bound by a given Transcription Factor to regulate gene expression or repression. In turn, DNA structured motifs are a mathematical counterpart that models sets of TFBSs that work in concert in the gene regulations processes of higher eukaryotic organisms. Typically, a structured motif is composed of an ordered set of isolated (or simple) motifs, separated by a variable, but somewhat constrained number of "irrelevant" base-pairs. Discovering structured motifs in a set of DNA sequences is a computationally hard problem that has been addressed by a number of authors using either a direct approach, or via the preliminary identification and successive combination of simple motifs.
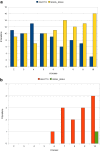
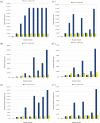
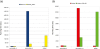
Results: We describe a computational tool, named SISMA, for the de-novo discovery of structured motifs in a set of DNA sequences. SISMA is an exact, enumerative algorithm, meaning that it finds all the motifs conforming to the specifications. It does so in two stages: first it discovers all the possible component simple motifs, then combines them in a way that respects the given constraints. We developed SISMA mainly with the aim of understanding the potential benefits of such a 2-stage approach w.r.t. direct methods. In fact, no 2-stage software was available for the general problem of structured motif discovery, but only a few tools that solved restricted versions of the problem. We evaluated SISMA against other published tools on a comprehensive benchmark made of both synthetic and real biological datasets. In a significant number of cases, SISMA outperformed the competitors, exhibiting a good performance also in most of the cases in which it was inferior.
Conclusions: A reflection on the results obtained lead us to conclude that a 2-stage approach can be implemented with many advantages over direct approaches. Some of these have to do with greater modularity, ease of parallelization, and the possibility to perform adaptive searches of structured motifs. As another consideration, we noted that most hard instances for SISMA were easy to detect in advance. In these cases one may initially opt for a direct method; or, as a viable alternative in most laboratories, one could run both direct and 2-stage tools in parallel, halting the computations when the first halts.
Figures


References
-
- Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R. Molecular Biology of the Gene. 6/e: Pearson International Edition; 2007.
-
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Rev Genet. 2007;8:206–216. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

