Examining histone posttranslational modification patterns by high-resolution mass spectrometry
- PMID: 22910200
- PMCID: PMC4116684
- DOI: 10.1016/B978-0-12-391940-3.00001-9
Examining histone posttranslational modification patterns by high-resolution mass spectrometry
Abstract
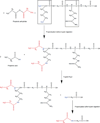
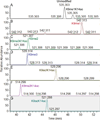
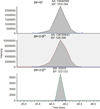
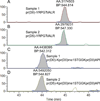
Histone variants and posttranslational modifications (PTMs) are essential for epigenetic regulation of transcriptional expression. Single and/or combinatorial PTMs of histones play important roles in development and disease formation. Mass spectrometry (MS) has been a powerful tool to study histone variants and PTMs, as it not only can identify novel PTMs but also can provide quantitative measurement of a spectrum of histone variants and PTMs in the same sample. In this chapter, we employ a combination of chemical derivation and high-resolution MS to identify and quantify multiple histone variants and PTMs. Histones are acid extracted and modified with propionyl groups and subsequently produces suitable sizes of fragments for MS analysis by trypsin digestion. The newly generated N-termini of histone peptides can be differentially marked by stable isotope labeling in a second reaction of propionylation, which enables direct comparison between two different samples in the following MS analysis.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




References
-
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. - PubMed
-
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. - PubMed
-
- Fuchs SM, Strahl BD. Antibody recognition of histone post-translational modifications: emerging issues and future prospects. Epigenomics. 2011;3:247–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

