Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model
- PMID: 22911607
- PMCID: PMC3406104
- DOI: 10.1371/journal.ppat.1002813
Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model
Abstract
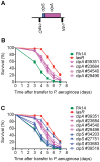
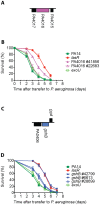
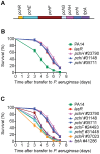
Pseudomonas aeruginosa strain PA14 is an opportunistic human pathogen capable of infecting a wide range of organisms including the nematode Caenorhabditis elegans. We used a non-redundant transposon mutant library consisting of 5,850 clones corresponding to 75% of the total and approximately 80% of the non-essential PA14 ORFs to carry out a genome-wide screen for attenuation of PA14 virulence in C. elegans. We defined a functionally diverse 180 mutant set (representing 170 unique genes) necessary for normal levels of virulence that included both known and novel virulence factors. Seven previously uncharacterized virulence genes (ABC transporters PchH and PchI, aminopeptidase PepP, ATPase/molecular chaperone ClpA, cold shock domain protein PA0456, putative enoyl-CoA hydratase/isomerase PA0745, and putative transcriptional regulator PA14_27700) were characterized with respect to pigment production and motility and all but one of these mutants exhibited pleiotropic defects in addition to their avirulent phenotype. We examined the collection of genes required for normal levels of PA14 virulence with respect to occurrence in P. aeruginosa strain-specific genomic regions, location on putative and known genomic islands, and phylogenetic distribution across prokaryotes. Genes predominantly contributing to virulence in C. elegans showed neither a bias for strain-specific regions of the P. aeruginosa genome nor for putatively horizontally transferred genomic islands. Instead, within the collection of virulence-related PA14 genes, there was an overrepresentation of genes with a broad phylogenetic distribution that also occur with high frequency in many prokaryotic clades, suggesting that in aggregate the genes required for PA14 virulence in C. elegans are biased towards evolutionarily conserved genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Moore NM, Flaws ML. Antimicrobial resistance mechanisms in Pseudomonas aeruginosa. Clin Lab Sci. 2011;24:47–51. - PubMed
-
- Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science. 2010;327:1010–1013. - PubMed
-
- Sintim HO, Smith JA, Wang J, Nakayama S, Yan L. Paradigm shift in discovering next-generation anti-infective agents: targeting quorum sensing, c-di-GMP signaling and biofilm formation in bacteria with small molecules. Future Med Chem. 2010;2:1005–1035. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

