C-terminal substitution of HBV core proteins with those from DHBV reveals that arginine-rich 167RRRSQSPRR175 domain is critical for HBV replication
- PMID: 22911745
- PMCID: PMC3401125
- DOI: 10.1371/journal.pone.0041087
C-terminal substitution of HBV core proteins with those from DHBV reveals that arginine-rich 167RRRSQSPRR175 domain is critical for HBV replication
Abstract
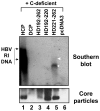
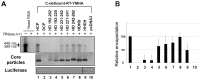
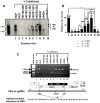
To investigate the contributions of carboxyl-terminal nucleic acid binding domain of HBV core (C) protein for hepatitis B virus (HBV) replication, chimeric HBV C proteins were generated by substituting varying lengths of the carboxyl-terminus of duck hepatitis B virus (DHBV) C protein for the corresponding regions of HBV C protein. All chimeric C proteins formed core particles. A chimeric C protein with 221-262 amino acids of DHBV C protein, in place of 146-185 amino acids of the HBV C protein, supported HBV pregenomic RNA (pgRNA) encapsidation and DNA synthesis: 40% amino acid sequence identity or 45% homology in the nucleic-acid binding domain of HBV C protein was sufficient for pgRNA encapsidation and DNA synthesis, although we predominantly detected spliced DNA. A chimeric C protein with 221-241 and 251-262 amino acids of DHBV C, in place of HBV C 146-166 and 176-185 amino acids, respectively, could rescue full-length DNA synthesis. However, a reciprocal C chimera with 242-250 of DHBV C ((242)RAGSPLPRS(250)) introduced in place of 167-175 of HBV C ((167)RRRSQSPRR(175)) significantly decreased pgRNA encapsidation and DNA synthesis, and full-length DNA was not detected, demonstrating that the arginine-rich (167)RRRSQSPRR(175) domain may be critical for efficient viral replication. Five amino acids differing between viral species (underlined above) were tested for replication rescue; R169 and R175 were found to be important.
Conflict of interest statement
Figures




Similar articles
-
Phosphoacceptors threonine 162 and serines 170 and 178 within the carboxyl-terminal RRRS/T motif of the hepatitis B virus core protein make multiple contributions to hepatitis B virus replication.J Virol. 2014 Aug;88(16):8754-67. doi: 10.1128/JVI.01343-14. Epub 2014 May 21. J Virol. 2014. PMID: 24850741 Free PMC article.
-
Sequences in the terminal protein and reverse transcriptase domains of the hepatitis B virus polymerase contribute to RNA binding and encapsidation.J Viral Hepat. 2014 Dec;21(12):882-93. doi: 10.1111/jvh.12225. Epub 2014 Jan 9. J Viral Hepat. 2014. PMID: 24401091 Free PMC article.
-
Regulation of multiple stages of hepadnavirus replication by the carboxyl-terminal domain of viral core protein in trans.J Virol. 2015 Mar;89(5):2918-30. doi: 10.1128/JVI.03116-14. Epub 2014 Dec 24. J Virol. 2015. PMID: 25540387 Free PMC article.
-
Phosphorylation of the Arginine-Rich C-Terminal Domains of the Hepatitis B Virus (HBV) Core Protein as a Fine Regulator of the Interaction between HBc and Nucleic Acid.Viruses. 2020 Jul 8;12(7):738. doi: 10.3390/v12070738. Viruses. 2020. PMID: 32650547 Free PMC article. Review.
-
Hepatitis B virus replication.World J Gastroenterol. 2007 Jan 7;13(1):48-64. doi: 10.3748/wjg.v13.i1.48. World J Gastroenterol. 2007. PMID: 17206754 Free PMC article. Review.
Cited by
-
An Alternatively Spliced Sirtuin 2 Isoform 5 Inhibits Hepatitis B Virus Replication from cccDNA by Repressing Epigenetic Modifications Made by Histone Lysine Methyltransferases.J Virol. 2020 Jul 30;94(16):e00926-20. doi: 10.1128/JVI.00926-20. Print 2020 Jul 30. J Virol. 2020. PMID: 32493816 Free PMC article.
-
Proteasomes regulate hepatitis B virus replication by degradation of viral core-related proteins in a two-step manner.Virus Genes. 2016 Oct;52(5):597-605. doi: 10.1007/s11262-016-1341-y. Epub 2016 Apr 22. Virus Genes. 2016. PMID: 27105855
-
Ca2+/Calmodulin-Dependent Protein Kinase II Inhibits Hepatitis B Virus Replication from cccDNA via AMPK Activation and AKT/mTOR Suppression.Microorganisms. 2022 Feb 23;10(3):498. doi: 10.3390/microorganisms10030498. Microorganisms. 2022. PMID: 35336076 Free PMC article.
-
PPIases Par14/Par17 Affect HBV Replication in Multiple Ways.Viruses. 2023 Feb 6;15(2):457. doi: 10.3390/v15020457. Viruses. 2023. PMID: 36851672 Free PMC article. Review.
-
Parvulin 14 and Parvulin 17 Bind to HBx and cccDNA and Upregulate Hepatitis B Virus Replication from cccDNA to Virion in an HBx-Dependent Manner.J Virol. 2019 Mar 5;93(6):e01840-18. doi: 10.1128/JVI.01840-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567987 Free PMC article.
References
-
- Brechot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56–S61. - PubMed
-
- Beames B, Lanford RE. Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology. 1993;194:597–607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

