Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures
- PMID: 22911969
- PMCID: PMC3428694
- DOI: 10.1128/mBio.00251-12
Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures
Abstract
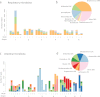
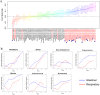
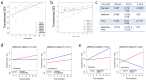
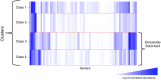
Pulmonary damage caused by chronic colonization of the cystic fibrosis (CF) lung by microbial communities is the proximal cause of respiratory failure. While there has been an effort to document the microbiome of the CF lung in pediatric and adult patients, little is known regarding the developing microflora in infants. We examined the respiratory and intestinal microbiota development in infants with CF from birth to 21 months. Distinct genera dominated in the gut compared to those in the respiratory tract, yet some bacteria overlapped, demonstrating a core microbiota dominated by Veillonella and Streptococcus. Bacterial diversity increased significantly over time, with evidence of more rapidly acquired diversity in the respiratory tract. There was a high degree of concordance between the bacteria that were increasing or decreasing over time in both compartments; in particular, a significant proportion (14/16 genera) increasing in the gut were also increasing in the respiratory tract. For 7 genera, gut colonization presages their appearance in the respiratory tract. Clustering analysis of respiratory samples indicated profiles of bacteria associated with breast-feeding, and for gut samples, introduction of solid foods even after adjustment for the time at which the sample was collected. Furthermore, changes in diet also result in altered respiratory microflora, suggesting a link between nutrition and development of microbial communities in the respiratory tract. Our findings suggest that nutritional factors and gut colonization patterns are determinants of the microbial development of respiratory tract microbiota in infants with CF and present opportunities for early intervention in CF with altered dietary or probiotic strategies.
Importance: While efforts have been focused on assessing the microbiome of pediatric and adult cystic fibrosis (CF) patients to understand how chronic colonization by these microbes contributes to pulmonary damage, little is known regarding the earliest development of respiratory and gut microflora in infants with CF. Our findings suggest that colonization of the respiratory tract by microbes is presaged by colonization of the gut and demonstrated a role of nutrition in development of the respiratory microflora. Thus, targeted dietary or probiotic strategies may be an effective means to change the course of the colonization of the CF lung and thereby improve patient outcomes.
Figures

Comment in
-
Exploring the parallel development of microbial systems in neonates with cystic fibrosis.mBio. 2012 Nov 6;3(6):e00408-12. doi: 10.1128/mBio.00408-12. mBio. 2012. PMID: 23131830 Free PMC article.
References
-
- Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. 2002. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr. Pulmonol. 34:91–100 - PubMed
-
- Bronstein MN, et al. 1992. Pancreatic insufficiency, growth, and nutrition in infants identified by newborn screening as having cystic fibrosis. J. Pediatr. 120:533–540 - PubMed
-
- Cox MJ, et al. 2010. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 5:e11044 http://dx.doi.org/10.1371/journal.pone.0011044 - PMC - PubMed
-
- Rosenecker J. 2000. Relations between the frequency of the deltaF508 mutation and the course of pulmonary disease in cystic fibrosis patients infected with Pseudomonas aeruginosa. Eur. J. Med. Res. 5:356–359 - PubMed
-
- McKone EF, Emerson SS, Edwards KL, Aitken ML. 2003. Effect of genotype on phenotype and mortality in cystic fibrosis: a retrospective cohort study. Lancet 361:1671–1676 - PubMed
Publication types
MeSH terms
Grants and funding
- U01 AT002388/AT/NCCIH NIH HHS/United States
- 1P20ES018175-02/ES/NIEHS NIH HHS/United States
- 5P20RR018787/RR/NCRR NIH HHS/United States
- P30 GM106394/GM/NIGMS NIH HHS/United States
- P42 ES007373/ES/NIEHS NIH HHS/United States
- P20RR16448/RR/NCRR NIH HHS/United States
- UH3 DK083993/DK/NIDDK NIH HHS/United States
- P20 GM103413/GM/NIGMS NIH HHS/United States
- R01 AI059694/AI/NIAID NIH HHS/United States
- R01 AI59694/AI/NIAID NIH HHS/United States
- P20 RR016448/RR/NCRR NIH HHS/United States
- P42 ES7373/ES/NIEHS NIH HHS/United States
- K24 AT003683/AT/NCCIH NIH HHS/United States
- P20RR016454/RR/NCRR NIH HHS/United States
- P20 RR018787/RR/NCRR NIH HHS/United States
- R01 HL074175/HL/NHLBI NIH HHS/United States
- P20-RR01878/RR/NCRR NIH HHS/United States
- 4UH3DK083993-02/DK/NIDDK NIH HHS/United States
- 8 P20 GM103413/GM/NIGMS NIH HHS/United States
- P20 RR016454/RR/NCRR NIH HHS/United States
- 2K24AT003683/AT/NCCIH NIH HHS/United States
- P20 ES018175/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

