Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method
- PMID: 22912385
- PMCID: PMC3482164
- DOI: 10.1161/CIRCRESAHA.112.273144
Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method
Abstract
Rationale: Cardiomyocytes (CMs) differentiated from human pluripotent stem cells (PSCs) are increasingly being used for cardiovascular research, including disease modeling, and hold promise for clinical applications. Current cardiac differentiation protocols exhibit variable success across different PSC lines and are primarily based on the application of growth factors. However, extracellular matrix is also fundamentally involved in cardiac development from the earliest morphogenetic events, such as gastrulation.
Objective: We sought to develop a more effective protocol for cardiac differentiation of human PSCs by using extracellular matrix in combination with growth factors known to promote cardiogenesis.
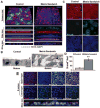
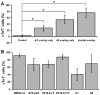
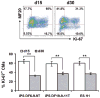
Methods and results: PSCs were cultured as monolayers on Matrigel, an extracellular matrix preparation, and subsequently overlayed with Matrigel. The matrix sandwich promoted an epithelial-to-mesenchymal transition as in gastrulation with the generation of N-cadherin-positive mesenchymal cells. Combining the matrix sandwich with sequential application of growth factors (Activin A, bone morphogenetic protein 4, and basic fibroblast growth factor) generated CMs with high purity (up to 98%) and yield (up to 11 CMs/input PSC) from multiple PSC lines. The resulting CMs progressively matured over 30 days in culture based on myofilament expression pattern and mitotic activity. Action potentials typical of embryonic nodal, atrial, and ventricular CMs were observed, and monolayers of electrically coupled CMs modeled cardiac tissue and basic arrhythmia mechanisms.
Conclusions: Dynamic extracellular matrix application promoted epithelial-mesenchymal transition of human PSCs and complemented growth factor signaling to enable robust cardiac differentiation.
Figures



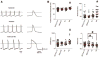

References
-
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. - PubMed
-
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a kdr+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. - PubMed
-
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and bmp signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. - PubMed
-
- Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, Skelton RJ, Ward-van Oostwaard D, Lim SM, Khammy O, Li X, Hawes SM, Davis RP, Goulburn AL, Passier R, Prall OW, Haynes JM, Pouton CW, Kaye DM, Mummery CL, Elefanty AG, Stanley EG. Nkx2-5(egfp/w) hescs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

