Intrinsic disorder in the human spliceosomal proteome
- PMID: 22912569
- PMCID: PMC3415423
- DOI: 10.1371/journal.pcbi.1002641
Intrinsic disorder in the human spliceosomal proteome
Abstract
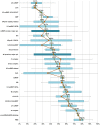
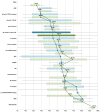
The spliceosome is a molecular machine that performs the excision of introns from eukaryotic pre-mRNAs. This macromolecular complex comprises in human cells five RNAs and over one hundred proteins. In recent years, many spliceosomal proteins have been found to exhibit intrinsic disorder, that is to lack stable native three-dimensional structure in solution. Building on the previous body of proteomic, structural and functional data, we have carried out a systematic bioinformatics analysis of intrinsic disorder in the proteome of the human spliceosome. We discovered that almost a half of the combined sequence of proteins abundant in the spliceosome is predicted to be intrinsically disordered, at least when the individual proteins are considered in isolation. The distribution of intrinsic order and disorder throughout the spliceosome is uneven, and is related to the various functions performed by the intrinsic disorder of the spliceosomal proteins in the complex. In particular, proteins involved in the secondary functions of the spliceosome, such as mRNA recognition, intron/exon definition and spliceosomal assembly and dynamics, are more disordered than proteins directly involved in assisting splicing catalysis. Conserved disordered regions in spliceosomal proteins are evolutionarily younger and less widespread than ordered domains of essential spliceosomal proteins at the core of the spliceosome, suggesting that disordered regions were added to a preexistent ordered functional core. Finally, the spliceosomal proteome contains a much higher amount of intrinsic disorder predicted to lack secondary structure than the proteome of the ribosome, another large RNP machine. This result agrees with the currently recognized different functions of proteins in these two complexes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, et al. (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375–387. - PubMed
-
- Zhou Z, Licklider LJ, Gygi SP, Reed R (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

