HSV-1 genome subnuclear positioning and associations with host-cell PML-NBs and centromeres regulate LAT locus transcription during latency in neurons
- PMID: 22912575
- PMCID: PMC3415458
- DOI: 10.1371/journal.ppat.1002852
HSV-1 genome subnuclear positioning and associations with host-cell PML-NBs and centromeres regulate LAT locus transcription during latency in neurons
Abstract
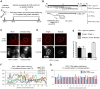
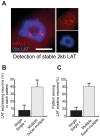
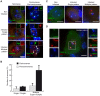
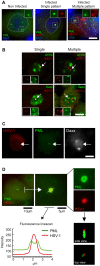
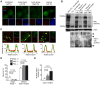
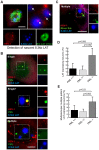
Major human pathologies are caused by nuclear replicative viruses establishing life-long latent infection in their host. During latency the genomes of these viruses are intimately interacting with the cell nucleus environment. A hallmark of herpes simplex virus type 1 (HSV-1) latency establishment is the shutdown of lytic genes expression and the concomitant induction of the latency associated (LAT) transcripts. Although the setting up and the maintenance of the latent genetic program is most likely dependent on a subtle interplay between viral and nuclear factors, this remains uninvestigated. Combining the use of in situ fluorescent-based approaches and high-resolution microscopic analysis, we show that HSV-1 genomes adopt specific nuclear patterns in sensory neurons of latently infected mice (28 days post-inoculation, d.p.i.). Latent HSV-1 genomes display two major patterns, called "Single" and "Multiple", which associate with centromeres, and with promyelocytic leukemia nuclear bodies (PML-NBs) as viral DNA-containing PML-NBs (DCP-NBs). 3D-image reconstruction of DCP-NBs shows that PML forms a shell around viral genomes and associated Daxx and ATRX, two PML partners within PML-NBs. During latency establishment (6 d.p.i.), infected mouse TGs display, at the level of the whole TG and in individual cells, a substantial increase of PML amount consistent with the interferon-mediated antiviral role of PML. "Single" and "Multiple" patterns are reminiscent of low and high-viral genome copy-containing neurons. We show that LAT expression is significantly favored within the "Multiple" pattern, which underlines a heterogeneity of LAT expression dependent on the viral genome copy number, pattern acquisition, and association with nuclear domains. Infection of PML-knockout mice demonstrates that PML/PML-NBs are involved in virus nuclear pattern acquisition, and negatively regulate the expression of the LAT. This study demonstrates that nuclear domains including PML-NBs and centromeres are functionally involved in the control of HSV-1 latency, and represent a key level of host/virus interaction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Roizman B, Knipe D, Whitley R (2007) Herpes Simplex Viruses. In: Fields Virology, 5th edition. Knipe D, Howley P, editors. Philadelphia: Lippincott Williams and Wilkins. 2501–2602.
-
- Knipe DM, Cliffe A (2008) Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6: 211–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

