HAL-2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors
- PMID: 22912597
- PMCID: PMC3415444
- DOI: 10.1371/journal.pgen.1002880
HAL-2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors
Abstract
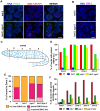
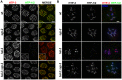
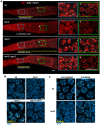
During meiosis, chromosomes align with their homologous pairing partners and stabilize this alignment through assembly of the synaptonemal complex (SC). Since the SC assembles cooperatively yet is indifferent to homology, pairing and SC assembly must be tightly coordinated. We identify HAL-2 as a key mediator in this coordination, showing that HAL-2 promotes pairing largely by preventing detrimental effects of SC precursors (SYP proteins). hal-2 mutants fail to establish pairing and lack multiple markers of chromosome movement mediated by pairing centers (PCs), chromosome sites that link chromosomes to cytoplasmic microtubules through nuclear envelope-spanning complexes. Moreover, SYP proteins load inappropriately along individual unpaired chromosomes in hal-2 mutants, and markers of PC-dependent movement and function are restored in hal-2; syp double mutants. These and other data indicate that SYP proteins can impede pairing and that HAL-2 promotes pairing predominantly but not exclusively by counteracting this inhibition, thereby enabling activation and regulation of PC function. HAL-2 concentrates in the germ cell nucleoplasm and colocalizes with SYP proteins in nuclear aggregates when SC assembly is prevented. We propose that HAL-2 functions to shepherd SYP proteins prior to licensing of SC assembly, preventing untimely interactions between SC precursors and chromosomes and allowing sufficient accumulation of precursors for rapid cooperative assembly upon homology verification.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Mlynarczyk-Evans S, Villeneuve AM (2010) Homologous chromosome pairing and synapsis during oogenesis. In: Verlhac M-H, Villeneuve AM, editors. Oogenesis: The Universal Process. Chichester, West Sussex, UK: Wiley-Blackwell. pp. 117–140.
-
- de Boer E, Heyting C (2006) The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115: 220–234. - PubMed
-
- Goldstein P (1987) Multiple synaptonemal complexes (polycomplexes): origin, structure and function. Cell Biol Int Rep 11: 759–796. - PubMed
-
- Jeffress JK, Page SL, Royer SK, Belden ED, Blumenstiel JP, et al. (2007) The formation of the central element of the synaptonemal complex may occur by multiple mechanisms: the roles of the N- and C-terminal domains of the Drosophila C(3)G protein in mediating synapsis and recombination. Genetics 177: 2445–2456. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

