Detection of parent-of-origin specific expression quantitative trait loci by cis-association analysis of gene expression in trios
- PMID: 22912676
- PMCID: PMC3422236
- DOI: 10.1371/journal.pone.0041695
Detection of parent-of-origin specific expression quantitative trait loci by cis-association analysis of gene expression in trios
Abstract
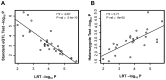
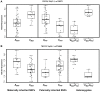
Parent-of-origin (PofO) effects, such as imprinting are a phenomenon in which homologous chromosomes exhibit differential gene expression and epigenetic modifications according to their parental origin. Such non-Mendelian inheritance patterns are generally ignored by conventional association studies, as these tests consider the maternal and paternal alleles as equivalent. To identify regulatory regions that show PofO effects on gene expression (imprinted expression Quantitative Trait Loci, ieQTLs), here we have developed a novel method in which we associate SNP genotypes of defined parental origin with gene expression levels. We applied this method to study 59 HapMap phase II parent-offspring trios. By analyzing mother/father/child trios, rules of Mendelian inheritance allowed the parental origin to be defined for ~95% of SNPs in each child. We used 680,475 informative SNPs and corresponding expression data for 92,167 probe sets from Affymetrix GeneChip Human Exon 1.0 ST arrays and performed four independent cis-association analyses with the expression level of RefSeq genes within 1 Mb using PLINK. Independent analyses of maternal and paternal genotypes identified two significant cis-ieQTLs (p<10(-7)) at which expression of genes SFT2D2 and SRRT associated exclusively with maternally inherited SNPs rs3753292 and rs6945374, respectively. 28 additional suggestive cis-associations with only maternal or paternal SNPs were found at a lower stringency threshold of p<10(-6), including associations with two known imprinted genes PEG10 and TRAPPC9, demonstrating the efficacy of our method. Furthermore, comparison of our method that utilizes independent analyses of maternal and paternal genotypes with the Likelihood Ratio Test (LRT) showed it to be more effective for detecting imprinting effects than the LRT. Our method represents a novel approach that can identify imprinted regulatory elements that control gene expression, suggesting novel PofO effects in the human genome.
Conflict of interest statement
Figures


References
-
- Gardner RJ, Mackay DJ, Mungall AJ, Polychronakos C, Siebert R, et al. (2000) An imprinted locus associated with transient neonatal diabetes mellitus. Hum Mol Genet 9: 589–596. - PubMed
-
- Reik W, Brown KW, Schneid H, Le Bouc Y, Bickmore W, et al. (1995) Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2-H19 domain. Hum Mol Genet 4: 2379–2385. - PubMed
-
- Morison IM, Ramsay JP, Spencer HG (2005) A census of mammalian imprinting. Trends Genet 21: 457–465. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

