Acquired inducible antimicrobial resistance in Gram-positive bacteria
- PMID: 22913355
- PMCID: PMC3464494
- DOI: 10.2217/fmb.12.63
Acquired inducible antimicrobial resistance in Gram-positive bacteria
Abstract
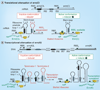
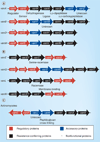
A major contributor to the emergence of antibiotic resistance in Gram-positive bacterial pathogens is the expansion of acquired, inducible genetic elements. Although acquired, inducible antibiotic resistance is not new, the interest in its molecular basis has been accelerated by the widening distribution and often 'silent' spread of the elements responsible, the diagnostic challenges of such resistance and the mounting limitations of available agents to treat Gram-positive infections. Acquired, inducible antibiotic resistance elements belong to the accessory genome of a species and are horizontally acquired by transformation/recombination or through the transfer of mobile DNA elements. The two key, but mechanistically very different, induction mechanisms are: ribosome-sensed induction, characteristic of the macrolide-lincosamide-streptogramin B antibiotics and tetracycline resistance, leading to ribosomal modifications or efflux pump activation; and resistance by cell surface-associated sensing of β-lactams (e.g., oxacillin), glycopeptides (e.g., vancomycin) and the polypeptide bacitracin, leading to drug inactivation or resistance due to cell wall alterations.
Conflict of interest statement
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Figures




References
-
-
Wozniak RAF, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010;8(8):552–563. ▪ This review of intergrative and conjugative elements discusses the core functions of these elements that allow for dynamic lateral gene flow, including regulation of conjugative transfer.
-
-
- Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 2011;35(5):856–871. - PubMed
-
- Rana SW, Kumar A, Walia SK, Berven K, Cumper K. Isolation of Tn 1546-like elements in vancomycin-resistant Enterococcus faecium isolated from wood frogs: an emerging risk for zoonotic bacterial infections to humans. J. Appl. Microbiol. 2011;110(1):35–43. - PubMed
Websites
-
- Marilyn C. Roberts, Ph.D. http://faculty.washington.edu/marilynr.
-
- Currently identified SCCmec types in S. aureus strains. www.sccmec.org/Pages/SCC_TypesEN.html.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
