Evolutionary genomics of host-use in bifurcating demes of RNA virus phi-6
- PMID: 22913547
- PMCID: PMC3495861
- DOI: 10.1186/1471-2148-12-153
Evolutionary genomics of host-use in bifurcating demes of RNA virus phi-6
Abstract
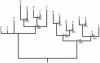
Background: Viruses are exceedingly diverse in their evolved strategies to manipulate hosts for viral replication. However, despite these differences, most virus populations will occasionally experience two commonly-encountered challenges: growth in variable host environments, and growth under fluctuating population sizes. We used the segmented RNA bacteriophage ϕ6 as a model for studying the evolutionary genomics of virus adaptation in the face of host switches and parametrically varying population sizes. To do so, we created a bifurcating deme structure that reflected lineage splitting in natural populations, allowing us to test whether phylogenetic algorithms could accurately resolve this 'known phylogeny'. The resulting tree yielded 32 clones at the tips and internal nodes; these strains were fully sequenced and measured for phenotypic changes in selected traits (fitness on original and novel hosts).
Results: We observed that RNA segment size was negatively correlated with the extent of molecular change in the imposed treatments; molecular substitutions tended to cluster on the Small and Medium RNA chromosomes of the virus, and not on the Large segment. Our study yielded a very large molecular and phenotypic dataset, fostering possible inferences on genotype-phenotype associations. Using further experimental evolution, we confirmed an inference on the unanticipated role of an allelic switch in a viral assembly protein, which governed viral performance across host environments.
Conclusions: Our study demonstrated that varying complexities can be simultaneously incorporated into experimental evolution, to examine the combined effects of population size, and adaptation in novel environments. The imposed bifurcating structure revealed that some methods for phylogenetic reconstruction failed to resolve the true phylogeny, owing to a paucity of molecular substitutions separating the RNA viruses that evolved in our study.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

