A framework for assessing the risk of resistance for anti-malarials in development
- PMID: 22913649
- PMCID: PMC3478971
- DOI: 10.1186/1475-2875-11-292
A framework for assessing the risk of resistance for anti-malarials in development
Abstract
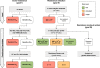
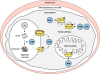
Resistance is a constant challenge for anti-infective drug development. Since they kill sensitive organisms, anti-infective agents are bound to exert an evolutionary pressure toward the emergence and spread of resistance mechanisms, if such resistance can arise by stochastic mutation events. New classes of medicines under development must be designed or selected to stay ahead in this vicious circle of resistance control. This involves both circumventing existing resistance mechanisms and selecting molecules which are resilient against the development and spread of resistance. Cell-based screening methods have led to a renaissance of new classes of anti-malarial medicines, offering us the potential to select and modify molecules based on their resistance potential. To that end, a standardized in vitro methodology to assess quantitatively these characteristics in Plasmodium falciparum during the early phases of the drug development process has been developed and is presented here. It allows the identification of anti-malarial compounds with overt resistance risks and the prioritization of the most robust ones. The integration of this strategy in later stages of development, registration, and deployment is also discussed.
Figures


References
-
- World Health Organization. Global report on antimalarial drug efficacy and drug resistance: 2000–2010. WHO, Geneva; 2010. p. 121.
-
- Looareesuwan S, Viravan C, Webster HK, Kyle DE, Hutchinson DB, Canfield CJ. Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. AmJTrop Med Hyg. 1996;54:62. - PubMed
-
- Korsinczky M, Chen N, Kotecka B, Saul A, Rieckmann K, Cheng Q. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother. 2000;44:2100–2108. doi: 10.1128/AAC.44.8.2100-2108.2000. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

