Murine gammaherpesvirus 68 LANA acts on terminal repeat DNA to mediate episome persistence
- PMID: 22915819
- PMCID: PMC3486315
- DOI: 10.1128/JVI.01656-12
Murine gammaherpesvirus 68 LANA acts on terminal repeat DNA to mediate episome persistence
Abstract
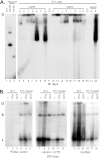
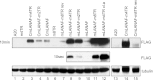
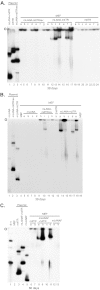
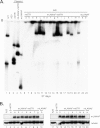
Murine gammaherpesvirus 68 (MHV68) ORF73 (mLANA) has sequence homology to Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen (LANA). LANA acts on the KSHV terminal repeat (TR) elements to mediate KSHV episome maintenance. Disruption of mLANA expression severely reduces the ability of MHV68 to establish latent infection in mice, consistent with the possibility that mLANA mediates episome persistence. Here we assess the roles of mLANA and MHV68 TR (mTR) elements in episome persistence. mTR-associated DNA persisted as an episome in latently MHV68-infected tumor cells, demonstrating that the mTR elements can serve as a cis-acting element for MHV68 episome maintenance. In some cases, both control vector and mTR-associated DNAs integrated into MHV68 episomal genomes. Therefore, we also assessed the roles of mTRs as well as mLANA in the absence of infection. DNA containing both mLANA and mTRs in cis persisted as an episome in murine A20 or MEF cells. In contrast, mTR DNA never persisted as an episome in the absence of mLANA. mLANA levels were increased when mLANA was expressed from its native promoters, and episome maintenance was more efficient with higher mLANA levels. Increased numbers of mTRs conferred more efficient episome maintenance, since DNA containing mLANA and eight mTR elements persisted more efficiently in A20 cells than did DNA with mLANA and two or four mTRs. Similar to KSHV LANA, mLANA broadly associated with mitotic chromosomes but relocalized to concentrated dots in the presence of episomes. Therefore, mLANA acts on mTR elements to mediate MHV68 episome persistence.
Figures



Similar articles
-
In Vivo Persistence of Chimeric Virus after Substitution of the Kaposi's Sarcoma-Associated Herpesvirus LANA DNA Binding Domain with That of Murid Herpesvirus 4.J Virol. 2018 Oct 12;92(21):e01251-18. doi: 10.1128/JVI.01251-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111565 Free PMC article.
-
Cross-species conservation of episome maintenance provides a basis for in vivo investigation of Kaposi's sarcoma herpesvirus LANA.PLoS Pathog. 2017 Sep 14;13(9):e1006555. doi: 10.1371/journal.ppat.1006555. eCollection 2017 Sep. PLoS Pathog. 2017. PMID: 28910389 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
Cited by
-
Latency-Associated Nuclear Antigen E3 Ubiquitin Ligase Activity Impacts Gammaherpesvirus-Driven Germinal Center B Cell Proliferation.J Virol. 2016 Aug 12;90(17):7667-83. doi: 10.1128/JVI.00813-16. Print 2016 Sep 1. J Virol. 2016. PMID: 27307564 Free PMC article.
-
Conditional mutagenesis in vivo reveals cell type- and infection stage-specific requirements for LANA in chronic MHV68 infection.PLoS Pathog. 2018 Jan 24;14(1):e1006865. doi: 10.1371/journal.ppat.1006865. eCollection 2018 Jan. PLoS Pathog. 2018. PMID: 29364981 Free PMC article.
-
Interplay of Murine Gammaherpesvirus 68 with NF-kappaB Signaling of the Host.Front Microbiol. 2016 Aug 17;7:1202. doi: 10.3389/fmicb.2016.01202. eCollection 2016. Front Microbiol. 2016. PMID: 27582728 Free PMC article. Review.
-
Kaposi's sarcoma herpesvirus exploits the DNA damage response to circularize its genome.Nucleic Acids Res. 2024 Feb 28;52(4):1814-1829. doi: 10.1093/nar/gkad1224. Nucleic Acids Res. 2024. PMID: 38180827 Free PMC article.
-
Kaposi's Sarcoma Herpesvirus Genome Persistence.Front Microbiol. 2016 Aug 12;7:1149. doi: 10.3389/fmicb.2016.01149. eCollection 2016. Front Microbiol. 2016. PMID: 27570517 Free PMC article. Review.
References
-
- Ballestas ME, Chatis PA, Kaye KM. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641–644 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

