Experimental relocation of the mitochondrial ATP9 gene to the nucleus reveals forces underlying mitochondrial genome evolution
- PMID: 22916027
- PMCID: PMC3420929
- DOI: 10.1371/journal.pgen.1002876
Experimental relocation of the mitochondrial ATP9 gene to the nucleus reveals forces underlying mitochondrial genome evolution
Abstract
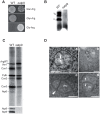
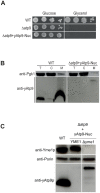
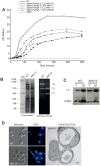
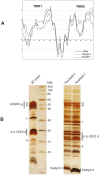
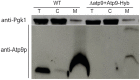
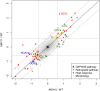
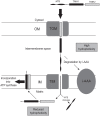
Only a few genes remain in the mitochondrial genome retained by every eukaryotic organism that carry out essential functions and are implicated in severe diseases. Experimentally relocating these few genes to the nucleus therefore has both therapeutic and evolutionary implications. Numerous unproductive attempts have been made to do so, with a total of only 5 successes across all organisms. We have taken a novel approach to relocating mitochondrial genes that utilizes naturally nuclear versions from other organisms. We demonstrate this approach on subunit 9/c of ATP synthase, successfully relocating this gene for the first time in any organism by expressing the ATP9 genes from Podospora anserina in Saccharomyces cerevisiae. This study substantiates the role of protein structure in mitochondrial gene transfer: expression of chimeric constructs reveals that the P. anserina proteins can be correctly imported into mitochondria due to reduced hydrophobicity of the first transmembrane segment. Nuclear expression of ATP9, while permitting almost fully functional oxidative phosphorylation, perturbs many cellular properties, including cellular morphology, and activates the heat shock response. Altogether, our study establishes a novel strategy for allotopic expression of mitochondrial genes, demonstrates the complex adaptations required to relocate ATP9, and indicates a reason that this gene was only transferred to the nucleus during the evolution of multicellular organisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283: 1476–1481. - PubMed
-
- Déquard-Chablat M, Sellem CH, Golik P, Bidard F, Martos A, et al. (2011) Two Nuclear Life Cycle-Regulated Genes Encode Interchangeable Subunits c of Mitochondrial ATP Synthase in Podospora anserina. Mol Biol Evol 28: 2063–2075. - PubMed
-
- Palmer JD (1997) Organelle genomes: going, going, gone!. Science 275: 790–791. - PubMed
-
- Claros MG, Perea J, Shu Y, Samatey FA, Popot JL, et al. (1995) Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur J Biochem 228: 762–771. - PubMed
-
- Allen JF (1993) Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J Theor Biol 165: 609–631. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

