Rho GTPase expression in human myeloid cells
- PMID: 22916134
- PMCID: PMC3420873
- DOI: 10.1371/journal.pone.0042563
Rho GTPase expression in human myeloid cells
Abstract
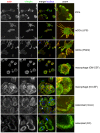
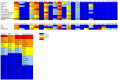
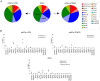
Myeloid cells are critical for innate immunity and the initiation of adaptive immunity. Strict regulation of the adhesive and migratory behavior is essential for proper functioning of these cells. Rho GTPases are important regulators of adhesion and migration; however, it is unknown which Rho GTPases are expressed in different myeloid cells. Here, we use a qPCR-based approach to investigate Rho GTPase expression in myeloid cells.We found that the mRNAs encoding Cdc42, RhoQ, Rac1, Rac2, RhoA and RhoC are the most abundant. In addition, RhoG, RhoB, RhoF and RhoV are expressed at low levels or only in specific cell types. More differentiated cells along the monocyte-lineage display lower levels of Cdc42 and RhoV, while RhoC mRNA is more abundant. In addition, the Rho GTPase expression profile changes during dendritic cell maturation with Rac1 being upregulated and Rac2 downregulated. Finally, GM-CSF stimulation, during macrophage and osteoclast differentiation, leads to high expression of Rac2, while M-CSF induces high levels of RhoA, showing that these cytokines induce a distinct pattern. Our data uncover cell type specific modulation of the Rho GTPase expression profile in hematopoietic stem cells and in more differentiated cells of the myeloid lineage.
Conflict of interest statement
Figures





References
-
- Banchereau J, Steinman RM (1998) Dendritic cells and the control of immunity. Nature 392: 245–252. - PubMed
-
- Moon SY, Zheng Y (2003) Rho GTPase-activating proteins in cell regulation. Trends Cell Biol 13: 13–22. - PubMed
-
- Rossman KL, Der CJ, Sondek J (2005) GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6: 167–180. - PubMed
-
- DerMardirossian C, Bokoch GM (2005) GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol 15: 356–363. - PubMed
-
- Dallery E, Galiegue-Zouitina S, Collyn-d'Hooghe M, Quief S, Denis C, et al. (1995) TTF, a gene encoding a novel small G protein, fuses to the lymphoma-associated LAZ3 gene by t(3;4) chromosomal translocation. Oncogene 10: 2171–2178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

