Three decades of farmed escapees in the wild: a spatio-temporal analysis of Atlantic salmon population genetic structure throughout Norway
- PMID: 22916215
- PMCID: PMC3419752
- DOI: 10.1371/journal.pone.0043129
Three decades of farmed escapees in the wild: a spatio-temporal analysis of Atlantic salmon population genetic structure throughout Norway
Abstract
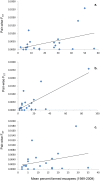
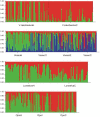
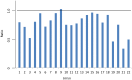
Each year, hundreds of thousands of domesticated farmed Atlantic salmon escape into the wild. In Norway, which is the world's largest commercial producer, many native Atlantic salmon populations have experienced large numbers of escapees on the spawning grounds for the past 15-30 years. In order to study the potential genetic impact, we conducted a spatio-temporal analysis of 3049 fish from 21 populations throughout Norway, sampled in the period 1970-2010. Based upon the analysis of 22 microsatellites, individual admixture, F(ST) and increased allelic richness revealed temporal genetic changes in six of the populations. These changes were highly significant in four of them. For example, 76% and 100% of the fish comprising the contemporary samples for the rivers Vosso and Opo were excluded from their respective historical samples at P=0.001. Based upon several genetic parameters, including simulations, genetic drift was excluded as the primary cause of the observed genetic changes. In the remaining 15 populations, some of which had also been exposed to high numbers of escapees, clear genetic changes were not detected. Significant population genetic structuring was observed among the 21 populations in the historical (global F(ST) =0.038) and contemporary data sets (global F(ST) =0.030), although significantly reduced with time (P=0.008). This reduction was especially distinct when looking at the six populations displaying temporal changes (global F(ST) dropped from 0.058 to 0.039, P=0.006). We draw two main conclusions: 1. The majority of the historical population genetic structure throughout Norway still appears to be retained, suggesting a low to modest overall success of farmed escapees in the wild; 2. Genetic introgression of farmed escapees in native salmon populations has been strongly population-dependent, and it appears to be linked with the density of the native population.
Conflict of interest statement
Figures

References
-
- Garcia de Leaniz C, Fleming IA, Einum S, Verspoor E, Jordan WC, et al. (2007) A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biological Reviews 82: 173–211. - PubMed
-
- Taylor EB (1991) A review of local adaptation in salmonidae, with particular reference to Pacific and Atlantic salmon. Aquaculture 98: 185–207.
-
- Stabell OB (1984) Homing and olfaction in salmonids - a critical review with special reference to the Atlantic salmon. Biological Reviews of the Cambridge Philosophical Society 59: 333–388.
-
- Ståhl G (1987) Genetic population strcuture of Atlantic salmon. In: Ryman N, Utter F, editors. Population genetics and fishery management: University of Washington press, Seattle. 121–140.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

