Acetylcholine-binding protein in the hemolymph of the planorbid snail Biomphalaria glabrata is a pentagonal dodecahedron (60 subunits)
- PMID: 22916297
- PMCID: PMC3423370
- DOI: 10.1371/journal.pone.0043685
Acetylcholine-binding protein in the hemolymph of the planorbid snail Biomphalaria glabrata is a pentagonal dodecahedron (60 subunits)
Abstract
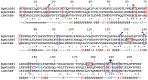
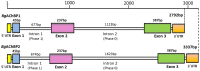
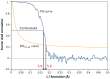
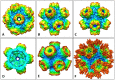
Nicotinic acetylcholine receptors (nAChR) play important neurophysiological roles and are of considerable medical relevance. They have been studied extensively, greatly facilitated by the gastropod acetylcholine-binding proteins (AChBP) which represent soluble structural and functional homologues of the ligand-binding domain of nAChR. All these proteins are ring-like pentamers. Here we report that AChBP exists in the hemolymph of the planorbid snail Biomphalaria glabrata (vector of the schistosomiasis parasite) as a regular pentagonal dodecahedron, 22 nm in diameter (12 pentamers, 60 active sites). We sequenced and recombinantly expressed two ∼25 kDa polypeptides (BgAChBP1 and BgAChBP2) with a specific active site, N-glycan site and disulfide bridge variation. We also provide the exon/intron structures. Recombinant BgAChBP1 formed pentamers and dodecahedra, recombinant BgAChBP2 formed pentamers and probably disulfide-bridged di-pentamers, but not dodecahedra. Three-dimensional electron cryo-microscopy (3D-EM) yielded a 3D reconstruction of the dodecahedron with a resolution of 6 Å. Homology models of the pentamers docked to the 6 Å structure revealed opportunities for chemical bonding at the inter-pentamer interfaces. Definition of the ligand-binding pocket and the gating C-loop in the 6 Å structure suggests that 3D-EM might lead to the identification of functional states in the BgAChBP dodecahedron.
Conflict of interest statement
Figures







References
-
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, et al. (2001) Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411: 269–276. - PubMed
-
- Celie PH, Klaassen RV, van Rossum-Fikkert SE, van Elk R, van Nierop P, et al. (2005) Crystal structure of acetylcholine-binding protein from Bulinus truncatus reveals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors. J Biol Chem 280: 26457–26466. - PubMed
-
- Lester HA, Dibas MI, Dahan DS, Leite JF, Dougherty DA (2004) Cys-loop receptors: new twists and turns. Trends Neurosci 27: 329–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

