Complexity in the wiring and regulation of plant circadian networks
- PMID: 22917516
- PMCID: PMC3427731
- DOI: 10.1016/j.cub.2012.07.025
Complexity in the wiring and regulation of plant circadian networks
Erratum in
- Curr Biol. 2013 Jan 7;23(1):95-6
Abstract
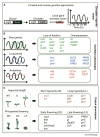
Endogenous circadian rhythms regulate many aspects of an organism's behavior, physiology and development. These daily oscillations synchronize with the environment to generate robust rhythms, resulting in enhanced fitness and growth vigor in plants. Collective studies over the years have focused on understanding the transcription-based oscillator in Arabidopsis. Recent advances combining mechanistic data with genome-wide approaches have contributed significantly to a more comprehensive understanding of the molecular interactions within the oscillator, and with clock-controlled pathways. This review focuses on the regulatory mechanisms within the oscillator, highlighting key connections between new and existing components, and direct mechanistic links to downstream pathways that control overt rhythms in the whole plant.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
From a repressilator-based circadian clock mechanism to an external coincidence model responsible for photoperiod and temperature control of plant architecture in Arabodopsis thaliana.Biosci Biotechnol Biochem. 2013;77(1):10-6. doi: 10.1271/bbb.120765. Epub 2013 Jan 7. Biosci Biotechnol Biochem. 2013. PMID: 23291766 Review.
-
Recent advances in understanding regulation of the Arabidopsis circadian clock by local cellular environment.F1000Res. 2020 Jan 27;9:F1000 Faculty Rev-51. doi: 10.12688/f1000research.21307.1. eCollection 2020. F1000Res. 2020. PMID: 32047621 Free PMC article. Review.
-
Molecular mechanisms at the core of the plant circadian oscillator.Nat Struct Mol Biol. 2016 Dec;23(12):1061-1069. doi: 10.1038/nsmb.3327. Epub 2016 Dec 6. Nat Struct Mol Biol. 2016. PMID: 27922614 Free PMC article. Review.
-
Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis.Plant Cell. 2014 Jan;26(1):79-87. doi: 10.1105/tpc.113.119842. Epub 2014 Jan 30. Plant Cell. 2014. PMID: 24481076 Free PMC article. Review.
-
A methyl transferase links the circadian clock to the regulation of alternative splicing.Nature. 2010 Nov 4;468(7320):112-6. doi: 10.1038/nature09470. Epub 2010 Oct 20. Nature. 2010. PMID: 20962777
Cited by
-
A global search for novel transcription factors impacting the Neurospora crassa circadian clock.G3 (Bethesda). 2021 Jun 17;11(6):jkab100. doi: 10.1093/g3journal/jkab100. G3 (Bethesda). 2021. PMID: 33792687 Free PMC article.
-
Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis.Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):E4802-10. doi: 10.1073/pnas.1513609112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261339 Free PMC article.
-
Evolutionary relationships among barley and Arabidopsis core circadian clock and clock-associated genes.J Mol Evol. 2015 Feb;80(2):108-19. doi: 10.1007/s00239-015-9665-0. Epub 2015 Jan 22. J Mol Evol. 2015. PMID: 25608480 Free PMC article.
-
Circadian clock regulation of mRNA translation through eukaryotic elongation factor eEF-2.Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):9605-10. doi: 10.1073/pnas.1525268113. Epub 2016 Aug 9. Proc Natl Acad Sci U S A. 2016. PMID: 27506798 Free PMC article.
-
Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors.Nat Commun. 2020 Oct 9;11(1):5089. doi: 10.1038/s41467-020-18832-8. Nat Commun. 2020. PMID: 33037196 Free PMC article.
References
-
- Yerushalmi S, Green RM. Evidence for the adaptive significance of circadian rhythms. Ecol Lett. 2009;12:970–981. - PubMed
-
- Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AAR. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30:333–349. - PubMed
-
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

