Why must T cells be cross-reactive?
- PMID: 22918468
- PMCID: PMC7097784
- DOI: 10.1038/nri3279
Why must T cells be cross-reactive?
Abstract
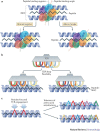
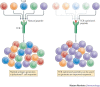
Clonal selection theory proposed that individual T cells are specific for a single peptide-MHC antigen. However, the repertoire of αβ T cell receptors (TCRs) is dwarfed by the vast array of potential foreign peptide-MHC complexes, and a comprehensive system requires each T cell to recognize numerous peptides and thus be cross-reactive. This compromise on specificity has profound implications because the chance of any natural peptide-MHC ligand being an optimal fit for its cognate TCR is small, as there will almost always be more-potent agonists. Furthermore, any TCR raised against a specific peptide-MHC complex in vivo can only be the best available solution from the naive T cell pool and is unlikely to be the best possible solution from the substantially greater number of TCRs that could theoretically be produced. This 'systems view' of TCR recognition provides a plausible cause for autoimmune disease and substantial scope for multiple therapeutic interventions.
Conflict of interest statement
The author declares no competing financial interests.
Figures



References
-
- Davis MM, et al. Ligand recognition by αβ T cell receptors. Annu. Rev. Immunol. 1998;16:523–544. - PubMed
-
- Davis MM. T cell receptor gene diversity and selection. Annu. Rev. Biochem. 1990;59:475–496. - PubMed
-
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. - PubMed
-
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

