Plant sphingolipid fatty acid 2-hydroxylases have unique characters unlike their animal and fungus counterparts
- PMID: 22918503
- PMCID: PMC3548854
- DOI: 10.4161/psb.21825
Plant sphingolipid fatty acid 2-hydroxylases have unique characters unlike their animal and fungus counterparts
Abstract
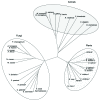
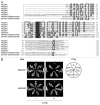
2-Hydroxy fatty acids mainly contained in sphingolipids are synthesized by a sphingolipid fatty acid 2-hydroxylase (FAH). Recently, two FAH homologs in Arabidopsis thaliana (AtFAH1 and AtFAH2), without any cytochrome b₅(Cb5)-like domains, which are essential for the function of Saccharomyces cerevisiae and mammalian FAH, were identified and both AtFAHs were shown to be activated by the interaction with Cb5. In this study, we compared FAHs of various plants, animals and fungi. Interestingly, only plants had two FAH homologs and none of plant FAHs had any Cb5-like domains. In addition, we showed from the interaction and expression analyses that AtFAHs interacted with multiple Cb5s probably in various tissues. Thus, plant FAHs may have evolved unlike animal and fungus FAHs.
Keywords: cytochrome b5; interaction; sphingolipid fatty acid 2-hydroxylase.
Figures


Similar articles
-
Arabidopsis sphingolipid fatty acid 2-hydroxylases (AtFAH1 and AtFAH2) are functionally differentiated in fatty acid 2-hydroxylation and stress responses.Plant Physiol. 2012 Jul;159(3):1138-48. doi: 10.1104/pp.112.199547. Epub 2012 May 25. Plant Physiol. 2012. PMID: 22635113 Free PMC article.
-
Functional association of cell death suppressor, Arabidopsis Bax inhibitor-1, with fatty acid 2-hydroxylation through cytochrome b₅.Plant J. 2009 Apr;58(1):122-34. doi: 10.1111/j.1365-313X.2008.03765.x. Epub 2009 Jan 19. Plant J. 2009. PMID: 19054355
-
Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids.J Biol Chem. 1997 Nov 7;272(45):28281-8. doi: 10.1074/jbc.272.45.28281. J Biol Chem. 1997. PMID: 9353282
-
Cytochrome b 5: A versatile electron carrier and regulator for plant metabolism.Front Plant Sci. 2022 Sep 23;13:984174. doi: 10.3389/fpls.2022.984174. eCollection 2022. Front Plant Sci. 2022. PMID: 36212330 Free PMC article. Review.
-
Fatty acid 2-Hydroxylation in mammalian sphingolipid biology.Biochim Biophys Acta. 2010 Apr;1801(4):405-14. doi: 10.1016/j.bbalip.2009.12.004. Epub 2009 Dec 21. Biochim Biophys Acta. 2010. PMID: 20026285 Free PMC article. Review.
Cited by
-
Fatty Acid 2-Hydroxylase and 2-Hydroxylated Sphingolipids: Metabolism and Function in Health and Diseases.Int J Mol Sci. 2023 Mar 3;24(5):4908. doi: 10.3390/ijms24054908. Int J Mol Sci. 2023. PMID: 36902339 Free PMC article. Review.
-
Sphingolipids with 2-hydroxy fatty acids aid in plasma membrane nanodomain organization and oxidative burst.Plant Physiol. 2022 Jun 1;189(2):839-857. doi: 10.1093/plphys/kiac134. Plant Physiol. 2022. PMID: 35312013 Free PMC article.
-
Plasma Membrane Microdomains Are Essential for Rac1-RbohB/H-Mediated Immunity in Rice.Plant Cell. 2016 Aug;28(8):1966-83. doi: 10.1105/tpc.16.00201. Epub 2016 Jul 27. Plant Cell. 2016. PMID: 27465023 Free PMC article.
-
Association of cytochrome b5 with ETR1 ethylene receptor signaling through RTE1 in Arabidopsis.Plant J. 2014 Feb;77(4):558-67. doi: 10.1111/tpj.12401. Plant J. 2014. PMID: 24635651 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
