DNA ligase I, the replicative DNA ligase
- PMID: 22918593
- PMCID: PMC3881551
- DOI: 10.1007/978-94-007-4572-8_17
DNA ligase I, the replicative DNA ligase
Abstract
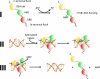
Multiple DNA ligation events are required to join the Okazaki fragments generated during lagging strand DNA synthesis. In eukaryotes, this is primarily carried out by members of the DNA ligase I family. The C-terminal catalytic region of these enzymes is composed of three domains: a DNA binding domain, an adenylation domain and an OB-fold domain. In the absence of DNA, these domains adopt an extended structure but transition into a compact ring structure when they engage a DNA nick, with each of the domains contacting the DNA. The non-catalytic N-terminal region of eukaryotic DNA ligase I is responsible for the specific participation of these enzymes in DNA replication. This proline-rich unstructured region contains the nuclear localization signal and a PCNA interaction motif that is critical for localization to replication foci and efficient joining of Okazaki fragments. DNA ligase I initially engages the PCNA trimer via this interaction motif which is located at the extreme N-terminus of this flexible region. It is likely that this facilitates an additional interaction between the DNA binding domain and the PCNA ring. The similar size and shape of the rings formed by the PCNA trimer and the DNA ligase I catalytic region when it engages a DNA nick suggest that these proteins interact to form a double-ring structure during the joining of Okazaki fragments. DNA ligase I also interacts with replication factor C, the factor that loads the PCNA trimeric ring onto DNA. This interaction, which is regulated by phosphorylation of the non-catalytic N-terminus of DNA ligase I, also appears to be critical for DNA replication.
Figures
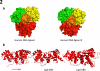
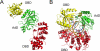

References
-
- Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. - PubMed
-
- Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet. 1996;13:489–491. - PubMed
-
- Bentley DJ, Harrison C, Ketchen AM, Redhead NJ, Samuel K, Waterfall M, Ansell JD, Melton DW. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J Cell Sci. 2002;115:1551–1561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

