Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p
- PMID: 22918946
- PMCID: PMC3459858
- DOI: 10.1091/mbc.E12-05-0400
Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p
Abstract
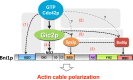
Actin filaments are dynamically reorganized to accommodate ever-changing cellular needs for intracellular transport, morphogenesis, and migration. Formins, a major family of actin nucleators, are believed to function as direct effectors of Rho GTPases, such as the polarity regulator Cdc42p. However, the presence of extensive redundancy has made it difficult to assess the in vivo significance of the low-affinity Rho GTPase-formin interaction and specifically whether Cdc42p polarizes the actin cytoskeleton via direct formin binding. Here we exploit a synthetically rewired budding yeast strain to eliminate the redundancy, making regulation of the formin Bni1p by Cdc42p essential for viability. Surprisingly, we find that direct Cdc42p-Bni1p interaction is dispensable for Bni1p regulation. Alternative paths linking Cdc42p and Bni1p via "polarisome" components Spa2p and Bud6p are also collectively dispensable. We identify a novel regulatory input to Bni1p acting through the Cdc42p effector, Gic2p. This pathway is sufficient to localize Bni1p to the sites of Cdc42p action and promotes a polarized actin organization in both rewired and wild-type contexts. We suggest that an indirect mechanism linking Rho GTPases and formins via Rho effectors may provide finer spatiotemporal control for the formin-nucleated actin cytoskeleton.
Figures







Similar articles
-
Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p).Mol Cell Biol. 2000 Sep;20(17):6244-58. doi: 10.1128/MCB.20.17.6244-6258.2000. Mol Cell Biol. 2000. PMID: 10938101 Free PMC article.
-
Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis.Science. 1997 Apr 4;276(5309):118-22. doi: 10.1126/science.276.5309.118. Science. 1997. PMID: 9082982
-
Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae.Mol Cell Biol. 2001 Feb;21(3):827-39. doi: 10.1128/MCB.21.3.827-839.2001. Mol Cell Biol. 2001. PMID: 11154270 Free PMC article.
-
Formins: signaling effectors for assembly and polarization of actin filaments.J Cell Sci. 2003 Jul 1;116(Pt 13):2603-11. doi: 10.1242/jcs.00611. J Cell Sci. 2003. PMID: 12775772 Review.
-
Formins as effector proteins of Rho GTPases.Small GTPases. 2014;5:e29513. doi: 10.4161/sgtp.29513. Epub 2014 Jun 10. Small GTPases. 2014. PMID: 24914801 Free PMC article. Review.
Cited by
-
The final cut: cell polarity meets cytokinesis at the bud neck in S. cerevisiae.Cell Mol Life Sci. 2016 Aug;73(16):3115-36. doi: 10.1007/s00018-016-2220-3. Epub 2016 Apr 16. Cell Mol Life Sci. 2016. PMID: 27085703 Free PMC article. Review.
-
Ratiometric GPCR signaling enables directional sensing in yeast.PLoS Biol. 2019 Oct 17;17(10):e3000484. doi: 10.1371/journal.pbio.3000484. eCollection 2019 Oct. PLoS Biol. 2019. PMID: 31622333 Free PMC article.
-
Temporal regulation of morphogenetic events in Saccharomyces cerevisiae.Mol Biol Cell. 2018 Aug 15;29(17):2069-2083. doi: 10.1091/mbc.E18-03-0188. Epub 2018 Jun 21. Mol Biol Cell. 2018. PMID: 29927361 Free PMC article.
-
Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis.Dev Cell. 2013 Jul 29;26(2):148-61. doi: 10.1016/j.devcel.2013.06.015. Dev Cell. 2013. PMID: 23906065 Free PMC article.
-
The Carboxy-Terminal Tails of Septins Cdc11 and Shs1 Recruit Myosin-II Binding Factor Bni5 to the Bud Neck in Saccharomyces cerevisiae.Genetics. 2015 Jul;200(3):843-62. doi: 10.1534/genetics.115.176503. Epub 2015 May 12. Genetics. 2015. PMID: 25971666 Free PMC article.
References
-
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

