ASGR1 and ASGR2, the Genes that Encode the Asialoglycoprotein Receptor (Ashwell Receptor), Are Expressed in Peripheral Blood Monocytes and Show Interindividual Differences in Transcript Profile
- PMID: 22919488
- PMCID: PMC3419429
- DOI: 10.1155/2012/283974
ASGR1 and ASGR2, the Genes that Encode the Asialoglycoprotein Receptor (Ashwell Receptor), Are Expressed in Peripheral Blood Monocytes and Show Interindividual Differences in Transcript Profile
Abstract
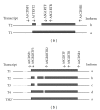
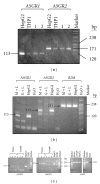
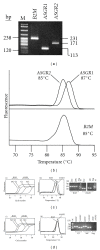
Background. The asialoglycoprotein receptor (ASGPR) is a hepatic receptor that mediates removal of potentially hazardous glycoconjugates from blood in health and disease. The receptor comprises two proteins, asialoglycoprotein receptor 1 and 2 (ASGR1 and ASGR2), encoded by the genes ASGR1 and ASGR2. Design and Methods. Using reverse transcription amplification (RT-PCR), expression of ASGR1 and ASGR2 was investigated in human peripheral blood monocytes. Results. Monocytes were found to express ASGR1 and ASGR2 transcripts. Correctly spliced transcript variants encoding different isoforms of ASGR1 and ASGR2 were present in monocytes. The profile of transcript variants from both ASGR1 and ASGR2 differed among individuals. Transcript expression levels were compared with the hepatocyte cell line HepG2 which produces high levels of ASGPR. Monocyte transcripts were 4 to 6 orders of magnitude less than in HepG2 but nonetheless readily detectable using standard RT-PCR. The monocyte cell line THP1 gave similar results to monocytes harvested from peripheral blood, indicating it may provide a suitable model system for studying ASGPR function in this cell type. Conclusions. Monocytes transcribe and correctly process transcripts encoding the constituent proteins of the ASGPR. Monocytes may therefore represent a mobile pool of the receptor, capable of reaching sites remote from the liver.
Figures
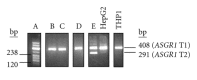

References
-
- Weigel PH, Yik JHN. Glycans as endocytosis signals: the cases of the asialoglycoprotein and hyaluronan/chondroitin sulfate receptors. Biochimica et Biophysica Acta. 2002;1572(2-3):341–363. - PubMed
-
- Ii M, Kurata H, Itoh N, Yamashina I, Kawasaki T. Molecular cloning and sequence analysis of cDNA encoding the macrophage lectin specific for galactose and N-acetylgalactosamine. The Journal of Biological Chemistry. 1990;265(19):11295–11298. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
