Polynucleotide phosphorylase has an impact on cell biology of Campylobacter jejuni
- PMID: 22919622
- PMCID: PMC3417634
- DOI: 10.3389/fcimb.2012.00030
Polynucleotide phosphorylase has an impact on cell biology of Campylobacter jejuni
Abstract
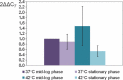
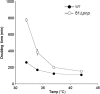
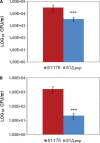
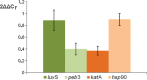
Polynucleotide phosphorylase (PNPase), encoded by the pnp gene, is known to degrade mRNA, mediating post-transcriptional regulation and may affect cellular functions. The role of PNPase is pleiotropic. As orthologs of the two major ribonucleases (RNase E and RNase II) of Escherichia coli are missing in the Campylobacter jejuni genome, in the current study the focus has been on the C. jejuni ortholog of PNPase. The effect of PNPase mutation on C. jejuni phenotypes and proteome was investigated. The inactivation of the pnp gene reduced significantly the ability of C. jejuni to adhere and to invade Ht-29 cells. Moreover, the pnp mutant strain exhibited a decrease in C. jejuni swimming ability and chick colonization. To explain effects of PNPase on C. jejuni 81-176 phenotype, the proteome of the pnp mutant and parental strains were compared. Overall, little variation in protein production was observed. Despite the predicted role of PNPase in mRNA regulation, the pnp mutation did not induce profound proteomic changes suggesting that other ribonucleases in C. jejuni might ensure this biological function in the absence of PNPase. Nevertheless, synthesis of proteins which are involved in virulence (LuxS, PEB3), motility (N-acetylneuraminic acid synthetase), stress-response (KatA, DnaK, Hsp90), and translation system (EF-Tu, EF-G) were modified in the pnp mutant strain suggesting a more specific role of PNPase in C. jejuni. In conclusion, PNPase deficiency induces limited but important consequences on C. jejuni biology that could explain swimming limitation, chick colonization delay, and the decrease of cell adhesion/invasion ability.
Keywords: 2D-electrophoresis; Campylobacter jejuni; chick colonization; in vitro virulence tests; polynucleotide phosphorylase.
Figures



Comment in
-
Exoribonucleases as modulators of virulence in pathogenic bacteria.Front Cell Infect Microbiol. 2012 May 11;2:65. doi: 10.3389/fcimb.2012.00065. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919656 Free PMC article. No abstract available.
Similar articles
-
Long-term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity.Appl Environ Microbiol. 2009 Dec;75(23):7310-8. doi: 10.1128/AEM.01366-09. Epub 2009 Oct 2. Appl Environ Microbiol. 2009. PMID: 19801468 Free PMC article.
-
The RNase R from Campylobacter jejuni has unique features and is involved in the first steps of infection.J Biol Chem. 2014 Oct 3;289(40):27814-24. doi: 10.1074/jbc.M114.561795. Epub 2014 Aug 6. J Biol Chem. 2014. PMID: 25100732 Free PMC article.
-
Transducer like proteins of Campylobacter jejuni 81-176: role in chemotaxis and colonization of the chicken gastrointestinal tract.Front Cell Infect Microbiol. 2015 May 27;5:46. doi: 10.3389/fcimb.2015.00046. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 26075188 Free PMC article.
-
Campylobacter jejuni: targeting host cells, adhesion, invasion, and survival.Appl Microbiol Biotechnol. 2023 May;107(9):2725-2754. doi: 10.1007/s00253-023-12456-w. Epub 2023 Mar 21. Appl Microbiol Biotechnol. 2023. PMID: 36941439 Free PMC article. Review.
-
Polyphosphate and associated enzymes as global regulators of stress response and virulence in Campylobacter jejuni.World J Gastroenterol. 2016 Sep 7;22(33):7402-14. doi: 10.3748/wjg.v22.i33.7402. World J Gastroenterol. 2016. PMID: 27672264 Free PMC article. Review.
Cited by
-
Adhesion, Biofilm Formation, and Genomic Features of Campylobacter jejuni Bf, an Atypical Strain Able to Grow under Aerobic Conditions.Front Microbiol. 2016 Jun 30;7:1002. doi: 10.3389/fmicb.2016.01002. eCollection 2016. Front Microbiol. 2016. PMID: 27446042 Free PMC article.
-
The impact of environmental conditions on Campylobacter jejuni survival in broiler faeces and litter.Infect Ecol Epidemiol. 2016 Jun 28;6:31685. doi: 10.3402/iee.v6.31685. eCollection 2016. Infect Ecol Epidemiol. 2016. PMID: 27357236 Free PMC article.
-
Insights into Campylobacter jejuni colonization and enteritis using a novel infant rabbit model.Sci Rep. 2016 Jun 30;6:28737. doi: 10.1038/srep28737. Sci Rep. 2016. PMID: 27357336 Free PMC article.
-
Identification of Ribonuclease Inhibitors for the Control of Pathogenic Bacteria.Int J Mol Sci. 2024 Jul 24;25(15):8048. doi: 10.3390/ijms25158048. Int J Mol Sci. 2024. PMID: 39125622 Free PMC article.
-
Next generation sequencing analysis reveals that the ribonucleases RNase II, RNase R and PNPase affect bacterial motility and biofilm formation in E. coli.BMC Genomics. 2015 Feb 14;16(1):72. doi: 10.1186/s12864-015-1237-6. BMC Genomics. 2015. PMID: 25757888 Free PMC article.
References
-
- Andersen M. T., Brøndsted L., Pearson B. M., Mulholland F., Parker M., Pin C., Wells J. M., Ingmer H. (2005). Diverse roles for HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology 151 905–915 - PubMed
-
- Andrade J. M., Pobre V., Silva I. J., Domingues S., Arraiano C. M. (2009). The role of 3′–5′ exoribonucleases in RNA degradation. Prog. Mol. Biol. Transl. Sci. 85187–229 - PubMed
-
- Arraiano C. M., Andrade J. M., Domingues S., Guinote I. B., Malecki M., Matos R. G., Moreira R. N., Pobre V., Reis F. P., Saramago M., Silva I. J., Viegas S. C. (2010). The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 34 883–923 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

