Comparative genomics of the Staphylococcus intermedius group of animal pathogens
- PMID: 22919635
- PMCID: PMC3417630
- DOI: 10.3389/fcimb.2012.00044
Comparative genomics of the Staphylococcus intermedius group of animal pathogens
Abstract
The Staphylococcus intermedius group consists of three closely related coagulase-positive bacterial species including S. intermedius, Staphylococcus pseudintermedius, and Staphylococcus delphini. S. pseudintermedius is a major skin pathogen of dogs, which occasionally causes severe zoonotic infections of humans. S. delphini has been isolated from an array of different animals including horses, mink, and pigeons, whereas S. intermedius has been isolated only from pigeons to date. Here we provide a detailed analysis of the S. pseudintermedius whole genome sequence in comparison to high quality draft S. intermedius and S. delphini genomes, and to other sequenced staphylococcal species. The core genome of the SIG was highly conserved with average nucleotide identity (ANI) between the three species of 93.61%, which is very close to the threshold of species delineation (95% ANI), highlighting the close-relatedness of the SIG species. However, considerable variation was identified in the content of mobile genetic elements, cell wall-associated proteins, and iron and sugar transporters, reflecting the distinct ecological niches inhabited. Of note, S. pseudintermedius ED99 contained a clustered regularly interspaced short palindromic repeat locus of the Nmeni subtype and S. intermedius contained both Nmeni and Mtube subtypes. In contrast to S. intermedius and S. delphini and most other staphylococci examined to date, S. pseudintermedius contained at least nine predicted reverse transcriptase Group II introns. Furthermore, S. pseudintermedius ED99 encoded several transposons which were largely responsible for its multi-resistant phenotype. Overall, the study highlights extensive differences in accessory genome content between closely related staphylococcal species inhabiting distinct host niches, providing new avenues for research into pathogenesis and bacterial host-adaptation.
Keywords: Staphylococcus; animal; antibiotic resistance; genomics; host-adaptation; pathogenesis.
Figures
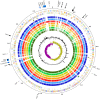
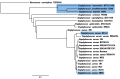


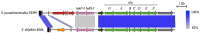
References
-
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., Formsma K., Gerdes S., Glass E. M., Kubal M., Meyer F., Olsen G. J., Olson R., Osterman A. L., Overbeek R. A., McNeil L. K., Paarmann D., Paczian T., Parrello B., Pusch G. D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75.10.1186/1471-2164-9-75 - DOI - PMC - PubMed
-
- Baba T., Kuwahara-Arai K., Uchiyama I., Takeuchi F., Ito T., Hiramatsu K. (2009). Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic Staphylococci. J. Bacteriol. 191, 1180–119010.1128/JB.00366-09 - DOI - PMC - PubMed
-
- Bannhoer J., Ben Zakour N. L., Waller A. S., Guardabassi L., Thoday K. M., van den Broek A. H. M., Fitzgerald J. R. (2007). Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J. Bacteriol. 189, 8685–869210.1128/JB.01150-07 - DOI - PMC - PubMed
-
- Bannoehr J., Ben Zakour N. L., Reglinski M., Inglis N. F., Prabhakaran S., Fossum E., Smith D. G., Wilson G. J., Cartwright R. A., Haas J., Hook M., van den Broek A. H., Thoday K. L., Fitzgerald J. R. (2011a). Genomic and surface proteomic analysis of the canine pathogen Staphylococcus pseudintermedius reveals proteins that mediate adherence to the extracellular matrix. Infect. Immun. 79, 3074–308610.1128/IAI.00137-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

