O-antigen and virulence profiling of shiga toxin-producing Escherichia coli by a rapid and cost-effective DNA microarray colorimetric method
- PMID: 22919652
- PMCID: PMC3417394
- DOI: 10.3389/fcimb.2012.00061
O-antigen and virulence profiling of shiga toxin-producing Escherichia coli by a rapid and cost-effective DNA microarray colorimetric method
Abstract
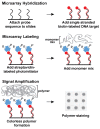
Shiga toxin-producing Escherichia coli (STEC) is a leading cause of foodborne illness worldwide. The present study developed the use of DNA microarrays with the ampliPHOX colorimetric method to rapidly detect and genotype STEC strains. A low-density 30-mer oligonucleotide DNA microarray was designed to target O-antigen gene clusters of 11 E. coli serogroups (O26, O45, O91, O103, O104, O111, O113, O121, O128, O145, and O157) that have been associated with the majority of STEC infections. In addition, the DNA microarray targeted 11 virulence genes, encoding adhesins, cytotoxins, proteases, and receptor proteins, which have been implicated in conferring increased ability to cause disease for STEC. Results from the validation experiments demonstrated that this microarray-based colorimetric method allowed for a rapid and accurate genotyping of STEC reference strains from environmental and clinical sources and from distinct geographical locations. Positive hybridization signals were detected only for probes targeting serotype and virulence genes known to be present in the STEC reference strains. Quantification analysis indicated that the mean pixel intensities of the signal for probes targeting O-antigen or virulence genes were at least three times higher when compared to the background. Furthermore, this microarray-based colorimetric method was then employed to genotype a group of E. coli isolates from watershed sediment and animal fecal samples that were collected from an important region for leafy-vegetable production in the central coast of California. The results indicated an accurate identification of O-type and virulence genes in the tested isolates and confirmed that the ampliPHOX colorimetric method with low-density DNA microarrays enabled a fast assessment of the virulence potential of STEC using low-cost reagents and instrumentation.
Keywords: DNA microarrays; Escherichia coli; STEC; Shiga toxin; ampliPHOX; foodborne pathogen; genotyping; photopolymerization.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

