Lamarckian evolution of the giant Mimivirus in allopatric laboratory culture on amoebae
- PMID: 22919682
- PMCID: PMC3417393
- DOI: 10.3389/fcimb.2012.00091
Lamarckian evolution of the giant Mimivirus in allopatric laboratory culture on amoebae
Abstract
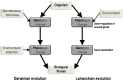
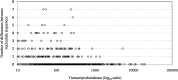
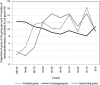
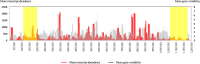
Acanthamoeba polyphaga Mimivirus has been subcultured 150 times on germ-free amoebae. This allopatric niche is very different from that found in the natural environment, where the virus is in competition with many other organisms. In this experiment, substantial gene variability and loss occurred concurrently with the emergence of phenotypically different viruses. We sought to quantify the respective roles of Lamarckian and Darwinian evolution during this experiment. We postulated that the Mimivirus genes that were down-regulated at the beginning of the allopatric laboratory culture and inactivated after 150 passages experienced Lamarckian evolution because phenotypic modifications preceded genotypic modifications, whereas we considered that genes that were highly transcribed in the new niche but were later inactivated obeyed Darwinian rules. We used the total transcript abundances and sequences described for the genes of Mimivirus at the beginning of its laboratory life and after 150 passages in allopatric culture on Acanthamoeba spp. We found a statistically significant positive correlation between the level of gene expression at the beginning of the culture and gene inactivation during the 150 passages. In particular, the mean transcript abundance at baseline was significantly lower for inactivated genes than for unchanged genes (165 ± 589 vs. 470 ± 1,625; p < 1e-3), and the mean transcript levels during the replication cycle of Mimivirus M1 were up to 8.5-fold lower for inactivated genes than for unchanged genes. In addition, proteins tended to be less frequently identified from purified virions in their early life in allopatric laboratory culture if they were encoded by variable genes than if they were encoded by conserved genes (9 vs. 15%; p = 0.062). Finally, Lamarckian evolution represented the evolutionary process encountered by 63% of the inactivated genes. Such observations may be explained by the lower level of DNA repair of useless genes.
Keywords: Darwinian evolution; Lamarckian evolution; Mimivirus; allopatry; gene expression profile; genome reduction; transcription profile.
Figures





Similar articles
-
Mimivirus: leading the way in the discovery of giant viruses of amoebae.Nat Rev Microbiol. 2017 Apr;15(4):243-254. doi: 10.1038/nrmicro.2016.197. Epub 2017 Feb 27. Nat Rev Microbiol. 2017. PMID: 28239153 Free PMC article. Review.
-
Analyses of the Kroon Virus Major Capsid Gene and Its Transcript Highlight a Distinct Pattern of Gene Evolution and Splicing among Mimiviruses.J Virol. 2018 Jan 2;92(2):e01782-17. doi: 10.1128/JVI.01782-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118120 Free PMC article.
-
Experimental Analysis of Mimivirus Translation Initiation Factor 4a Reveals Its Importance in Viral Protein Translation during Infection of Acanthamoeba polyphaga.J Virol. 2018 Apr 27;92(10):e00337-18. doi: 10.1128/JVI.00337-18. Print 2018 May 15. J Virol. 2018. PMID: 29514904 Free PMC article.
-
Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae.Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17486-91. doi: 10.1073/pnas.1110889108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987820 Free PMC article.
-
Mimivirus and its virophage.Annu Rev Genet. 2009;43:49-66. doi: 10.1146/annurev-genet-102108-134255. Annu Rev Genet. 2009. PMID: 19653859 Review.
Cited by
-
Modulation of the expression of mimivirus-encoded translation-related genes in response to nutrient availability during Acanthamoeba castellanii infection.Front Microbiol. 2015 Jun 1;6:539. doi: 10.3389/fmicb.2015.00539. eCollection 2015. Front Microbiol. 2015. PMID: 26082761 Free PMC article.
-
The number of genes encoding repeat domain-containing proteins positively correlates with genome size in amoebal giant viruses.Virus Evol. 2018 Jan 3;4(1):vex039. doi: 10.1093/ve/vex039. eCollection 2018 Jan. Virus Evol. 2018. PMID: 29308275 Free PMC article.
-
Mimivirus: leading the way in the discovery of giant viruses of amoebae.Nat Rev Microbiol. 2017 Apr;15(4):243-254. doi: 10.1038/nrmicro.2016.197. Epub 2017 Feb 27. Nat Rev Microbiol. 2017. PMID: 28239153 Free PMC article. Review.
-
Microbial genomics challenge Darwin.Front Cell Infect Microbiol. 2012 Oct 12;2:127. doi: 10.3389/fcimb.2012.00127. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 23091803 Free PMC article. No abstract available.
-
Acanthamoeba polyphaga mimivirus and other giant viruses: an open field to outstanding discoveries.Virol J. 2014 Jun 30;11:120. doi: 10.1186/1743-422X-11-120. Virol J. 2014. PMID: 24976356 Free PMC article. Review.
References
-
- Boyer M., Azza S., Barrassi L., Klose T., Campocasso A., Pagnier I., Fournous G., Borg A., Robert C., Zhang X., Desnues C., Henrissat B., Rossmann M. G., La S. B., Raoult D. (2011). Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. U.S.A. 108, 10296–10301 10.1073/pnas.1101118108 - DOI - PMC - PubMed
-
- Colson P., Yutin N., Shabalina S. A., Robert C., Fournous G., La Scola B., Raoult D., Koonin E. V. (2011). Viruses with more than 1, 000 genes: Mamavirus, a new Acanthamoeba polyphaga Mimivirus strain, and reannotation of Mimivirus genes. Genome Biol. Evol. 3, 737–742 10.1093/gbe/evr048 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

